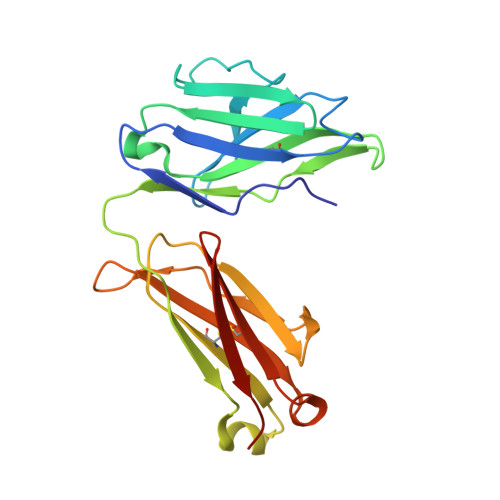
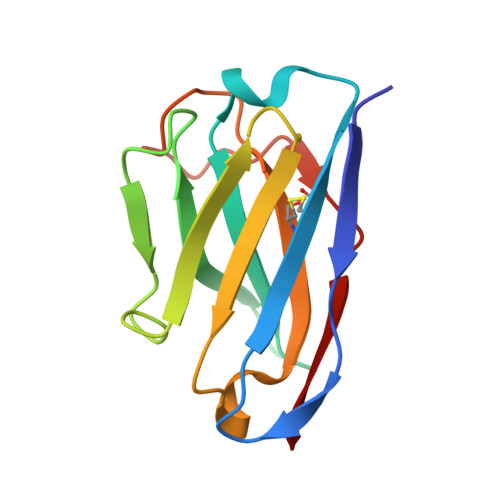
Nanobodies counteract the toxicity of an amyloidogenic light chain by stabilizing a partially open dimeric conformation.
Broggini, L., Barzago, M.M., Speranzini, V., Schulte, T., Sonzini, F., Giono, M., Romeo, M., Milani, P., Caminito, S., Mazzini, G., Rognoni, P., Merlini, G., Pappone, C., Anastasia, L., Nuvolone, M., Palladini, G., Diomede, L., Ricagno, S.(2023) J Mol Biology 435: 168320-168320
- PubMed: 37865287
- DOI: https://doi.org/10.1016/j.jmb.2023.168320
- Primary Citation of Related Structures:
8P88, 8P89 - PubMed Abstract:
Light chain amyloidosis (AL) is a systemic disease where fibrillar deposition of misfolded immunoglobulin light chains (LCs) severely affects organ function and results in poor prognosis for patients, especially when heart involvement is severe. Particularly relevant in this context is the cardiotoxicity exerted by still uncharacterized soluble LC species. Here, with the final goal of identifying alternative therapeutic strategies to tackle AL amyloidosis, we produced five llama-derived nanobodies (Nbs) specific against H3, a well-characterized amyloidogenic and cardiotoxic LC from an AL patient with severe cardiac involvement. We found that Nbs are specific and potent agents capable of abolishing H3 soluble toxicity in C. elegans in vivo model. Structural characterization of H3-Nb complexes revealed that the protective effect of Nbs is related to their ability to bind to the H3 V L domain and stabilise an unexpected partially open LC dimer in which the two V L domains no longer interact with each other. Thus, while identifying potent inhibitors of LC soluble toxicity, we also describe the first non-native structure of an amyloidogenic LC that may represent a crucial step in toxicity and aggregation mechanisms.
- Institute of Molecular and Translational Cardiology, IRCCS Policlinico San Donato, Piazza Malan 2, 20097 San Donato Milanese, Italy.
Organizational Affiliation: