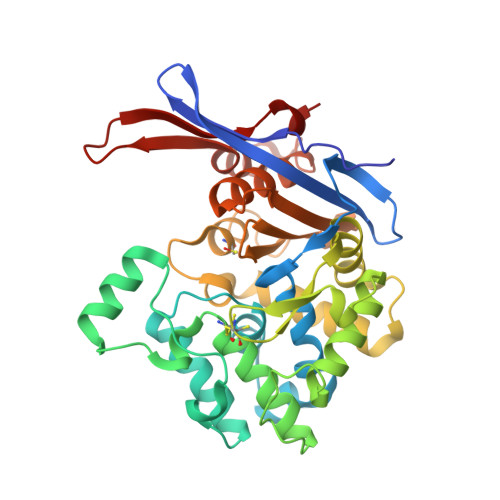
Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Loch, J.I., Worsztynowicz, P., Sliwiak, J., Grzechowiak, M., Imiolczyk, B., Pokrywka, K., Chwastyk, M., Gilski, M., Jaskolski, M.(2023) Acta Crystallogr D Struct Biol 79: 775-791
- PubMed: 37494066
- DOI: https://doi.org/10.1107/S2059798323005648
- Primary Citation of Related Structures:
8CLY, 8CLZ, 8COL, 8ORI, 8OSW - PubMed Abstract:
The genome of Rhizobium etli, a nitrogen-fixing bacterial symbiont of legume plants, encodes two L-asparaginases, ReAIV and ReAV, that have no similarity to the well characterized enzymes of class 1 (bacterial type) and class 2 (plant type). It has been hypothesized that ReAIV and ReAV might belong to the same structural class 3 despite their low level of sequence identity. When the crystal structure of the inducible and thermolabile protein ReAV was solved, this hypothesis gained a stronger footing because the key residues of ReAV are also present in the sequence of the constitutive and thermostable ReAIV protein. High-resolution crystal structures of ReAIV now confirm that it is a class 3 L-asparaginase that is structurally similar to ReAV but with important differences. The most striking differences concern the peculiar hydration patterns of the two proteins, the presence of three internal cavities in ReAIV and the behavior of the zinc-binding site. ReAIV has a high pH optimum (9-11) and a substrate affinity of ∼1.3 mM at pH 9.0. These parameters are not suitable for the direct application of ReAIV as an antileukemic drug, although its thermal stability and lack of glutaminase activity would be of considerable advantage. The five crystal structures of ReAIV presented in this work allow a possible enzymatic scenario to be postulated in which the zinc ion coordinated in the active site is a dispensable element. The catalytic nucleophile seems to be Ser47, which is part of two Ser-Lys tandems in the active site. The structures of ReAIV presented here may provide a basis for future enzyme-engineering experiments to improve the kinetic parameters for medicinal applications.
- Department of Crystal Chemistry and Crystal Physics, Faculty of Chemistry, Jagiellonian University, Krakow, Poland.
Organizational Affiliation: