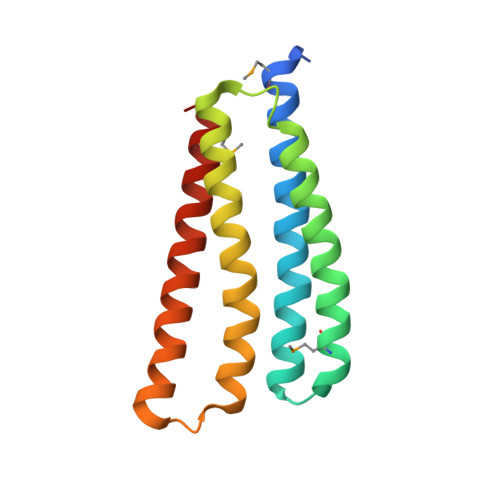
Rex1BD and the 14-3-3 protein control heterochromatin organization at tandem repeats by linking RNAi and HDAC.
Gao, J., Sun, W., Li, J., Ban, H., Zhang, T., Liao, J., Kim, N., Lee, S.H., Dong, Q., Madramootoo, R., Chen, Y., Li, F.(2023) Proc Natl Acad Sci U S A 120: e2309359120-e2309359120
- PubMed: 38048463
- DOI: https://doi.org/10.1073/pnas.2309359120
- Primary Citation of Related Structures:
8J0H - PubMed Abstract:
Tandem DNA repeats are often organized into heterochromatin that is crucial for genome organization and stability. Recent studies revealed that individual repeats within tandem DNA repeats can behave very differently. How DNA repeats are assembled into distinct heterochromatin structures remains poorly understood. Here, we developed a genome-wide genetic screen using a reporter gene at different units in a repeat array. This screen led to identification of a conserved protein Rex1BD required for heterochromatin silencing. Our structural analysis revealed that Rex1BD forms a four-helix bundle structure with a distinct charged electrostatic surface. Mechanistically, Rex1BD facilitates the recruitment of Clr6 histone deacetylase (HDAC) by interacting with histones. Interestingly, Rex1BD also interacts with the 14-3-3 protein Rad25, which is responsible for recruiting the RITS (RNA-induced transcriptional silencing) complex to DNA repeats. Our results suggest that coordinated action of Rex1BD and Rad25 mediates formation of distinct heterochromatin structure at DNA repeats via linking RNAi and HDAC pathways.
- Department of Biology, New York University, New York, NY 10003.
Organizational Affiliation: