Cryo-EM structure of hnRAC1-2I fibril.
Li, D.N., Ma, Y.Y., Li, D., Dai, B., Liu, C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Find similar proteins by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GLY-PHI-GLY-GLY-ASN-ASP-ASN-PHE-GLY | A [auth D], AA [auth 0], B [auth E], BA [auth 3], C [auth F], D [auth I], E [auth J], F [auth L], G [auth M], H [auth P], I [auth S], J [auth T], K [auth W], L [auth X], M [auth Z], N [auth a], O [auth c], P [auth d], Q [auth e], R [auth i], S [auth j], T [auth m], U [auth p], V [auth q], W [auth s], X [auth t], Y [auth w], Z [auth x] | 9 | Homo sapiens | Mutation(s): 0 | 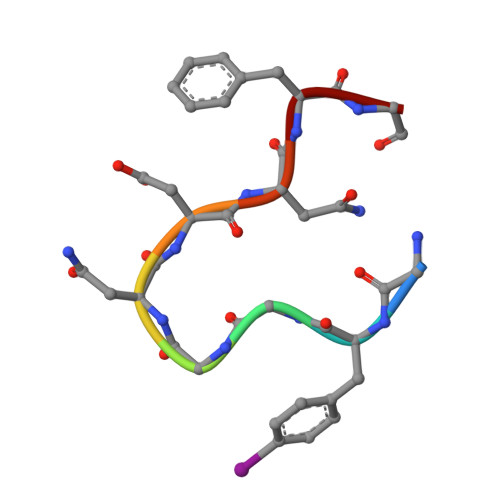 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P09651 (Homo sapiens) Explore P09651 Go to UniProtKB: P09651 | |||||
PHAROS: P09651 GTEx: ENSG00000135486 | |||||
Entity Groups | |||||
| UniProt Group | P09651 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PHI Query on PHI | A [auth D] AA [auth 0] B [auth E] BA [auth 3] C [auth F] A [auth D], AA [auth 0], B [auth E], BA [auth 3], C [auth F], D [auth I], E [auth J], F [auth L], G [auth M], H [auth P], I [auth S], J [auth T], K [auth W], L [auth X], M [auth Z], N [auth a], O [auth c], P [auth d], Q [auth e], R [auth i], S [auth j], T [auth m], U [auth p], V [auth q], W [auth s], X [auth t], Y [auth w], Z [auth x] | L-PEPTIDE LINKING | C9 H10 I N O2 |  | PHE |