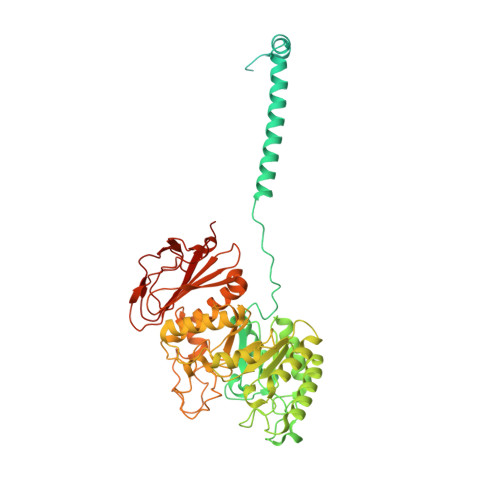
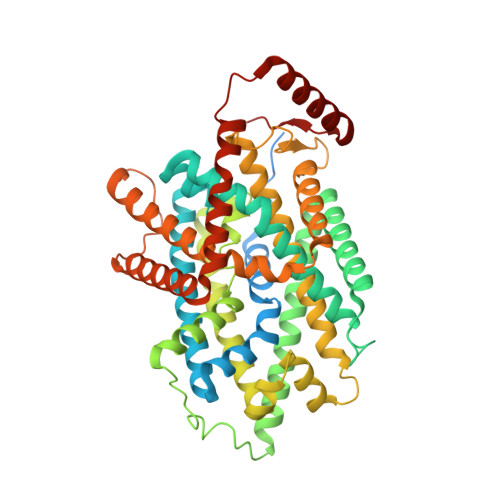
Structural insights into the substrate transport mechanism of the amino acid transporter complex.
Yang, H., Shi, T., Dong, J., Zhang, T., Li, Y., Guo, Y., Yuan, Y., Yang, L., Dong, J.T., Yan, R.(2025) J Biological Chem 301: 110569-110569
- PubMed: 40780412
- DOI: https://doi.org/10.1016/j.jbc.2025.110569
- Primary Citation of Related Structures:
8IDA, 8J8L, 8J8M, 8X0W - PubMed Abstract:
The L -type amino acid transporter 1 (LAT1), in complex with its ancillary protein 4F2hc, mediates the sodium-independent antiport of large neutral amino acids across the plasma membrane. LAT1 preferentially transports substrates, such as L -leucine, L -tyrosine, and L -tryptophan, thyroid hormones, and drugs like 3,4-dihydroxyphenylalanine. Its pivotal role in cancer development and progression has established LAT1 as a promising therapeutic target. While prior studies have resolved the LAT1-4F2hc architecture and inhibitor interactions, the molecular basis of LAT1 substrate selectivity remains elusive. Here, we present the cryo-EM structures of LAT1-4F2hc bound to L -tyrosine, L -tryptophan, L -leucine, and 3,4-dihydroxyphenylalanine, revealing distinct substrate binding modes. Comparative structural analysis highlights differences between LAT1 and LAT2 in substrate coordination, driven by key residues near the binding pocket that influence transport efficiency. These findings advance our mechanistic understanding of the LAT1-4F2hc complex and provide valuable insights for structure-based drug design targeting LAT1.
- Department of Biochemistry, SUSTech Homeostatic Medicine Institute, School of Medicine, Key University Laboratory of Metabolism and Health of Guangdong, Institute for Biological Electron Microscopy, Southern University of Science and Technology, Shenzhen, Guangdong, China.
Organizational Affiliation: