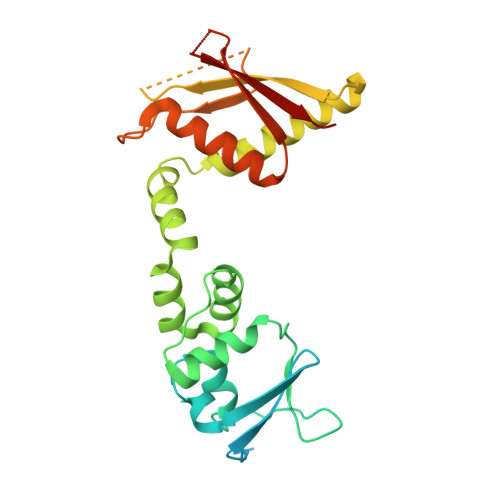
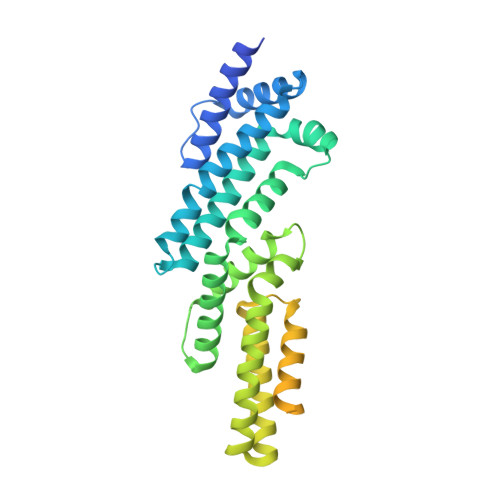
Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Jiang, W., Wang, W., Kong, Y., Zheng, S.(2023) Sci Adv 9: eadg8369-eadg8369
- PubMed: 37450587
- DOI: https://doi.org/10.1126/sciadv.adg8369
- Primary Citation of Related Structures:
8I79, 8JKB - PubMed Abstract:
G protein-coupled receptor (GPCR) signaling is precisely controlled to avoid overstimulation that results in detrimental consequences. Gβγ signaling is negatively regulated by a Cullin3 (Cul3)-dependent E3 ligase, KCTD5, which triggers ubiquitination and degradation of free Gβγ. Here, we report the cryo-electron microscopy structures of the KCTD5-Gβγ fusion complex and the KCTD7-Cul3 complex. KCTD5 in pentameric form engages symmetrically with five copies of Gβγ through its C-terminal domain. The unique pentameric assembly of the KCTD5/Cul3 E3 ligase places the ubiquitin-conjugating enzyme (E2) and the modification sites of Gβγ in close proximity and allows simultaneous transfer of ubiquitin from E2 to five Gβγ subunits. Moreover, we show that ubiquitination of Gβγ by KCTD5 is important for fine-tuning cyclic adenosine 3´,5´-monophosphate signaling of GPCRs. Our studies provide unprecedented insights into mechanisms of substrate recognition by unusual pentameric E3 ligases and highlight the KCTD family as emerging regulators of GPCR signaling.
- Graduate School of Peking Union Medical College, Beijing 100730, China.
Organizational Affiliation: