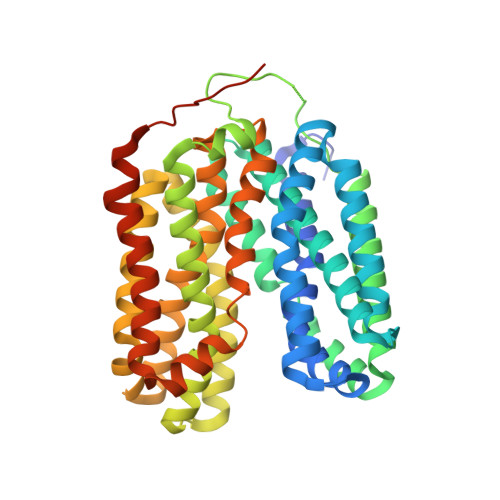
Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota.
Jaunet-Lahary, T., Shimamura, T., Hayashi, M., Nomura, N., Hirasawa, K., Shimizu, T., Yamashita, M., Tsutsumi, N., Suehiro, Y., Kojima, K., Sudo, Y., Tamura, T., Iwanari, H., Hamakubo, T., Iwata, S., Okazaki, K.I., Hirai, T., Yamashita, A.(2023) Nat Commun 14: 1730-1730
- PubMed: 37012268
- DOI: https://doi.org/10.1038/s41467-023-36883-5
- Primary Citation of Related Structures:
8HPJ, 8HPK - PubMed Abstract:
An oxalate-degrading bacterium in the gut microbiota absorbs food-derived oxalate to use this as a carbon and energy source, thereby reducing the risk of kidney stone formation in host animals. The bacterial oxalate transporter OxlT selectively uptakes oxalate from the gut to bacterial cells with a strict discrimination from other nutrient carboxylates. Here, we present crystal structures of oxalate-bound and ligand-free OxlT in two distinct conformations, occluded and outward-facing states. The ligand-binding pocket contains basic residues that form salt bridges with oxalate while preventing the conformational switch to the occluded state without an acidic substrate. The occluded pocket can accommodate oxalate but not larger dicarboxylates, such as metabolic intermediates. The permeation pathways from the pocket are completely blocked by extensive interdomain interactions, which can be opened solely by a flip of a single side chain neighbouring the substrate. This study shows the structural basis underlying metabolic interactions enabling favourable symbiosis.
- Research Center for Computational Science, Institute for Molecular Science, National Institutes of Natural Sciences, Okazaki, 444-8585, Japan.
Organizational Affiliation: