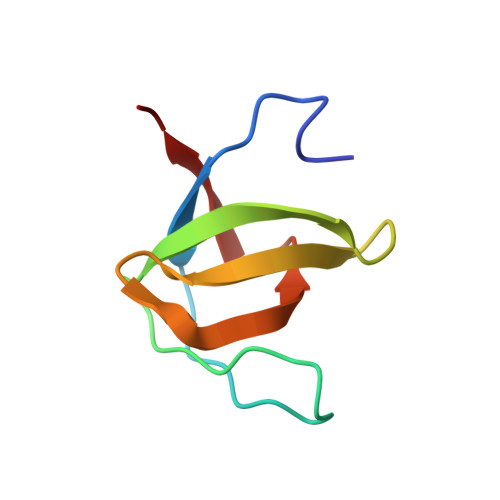
Crystal Structure of the SH3 Domain of ASAP1 in Complex with the Proline Rich Motif (PRM) of MICAL1 Reveals a Unique SH3/PRM Interaction Mode.
Jia, X., Lin, L., Xu, S., Li, L., Wei, Z., Yu, C., Niu, F.(2023) Int J Mol Sci 24
- PubMed: 36674928
- DOI: https://doi.org/10.3390/ijms24021414
- Primary Citation of Related Structures:
8HLO - PubMed Abstract:
SH3 domains are common protein binding modules. The target sequence of SH3 domains is usually a proline-rich motif (PRM) containing a minimal "PxxP" sequence. The mechanism of how different SH3 domains specifically choose their targets from vast PxxP-containing sequences is still not very clear, as many reported SH3/PRM interactions are weak and promiscuous. Here, we identified the binding of the SH3 domain of ASAP1 to the PRM of MICAL1 with a sub-μM binding affinity, and determined the crystal structure of ASAP1-SH3 and MICAL1-PRM complex. Our structural and biochemical analyses revealed that the target-binding pocket of ASAP1-SH3 contains two negatively charged patches to recognize the "xPx + Px+" sequence in MICAL1-PRM and consequently strengthen the interaction, differing from the typical SH3/PRM interaction. This unique PRM-binding pocket is also found in the SH3 domains of GTPase Regulator associated with focal adhesion kinase (GRAF) and Src kinase associated phosphoprotein 1 (SKAP1), which we named SH3 AGS . In addition, we searched the Swiss-Prot database and found ~130 proteins with the SH3 AGS -binding PRM in silico. Finally, gene ontology analysis suggests that the strong interaction between the SH3 AGS -containing proteins and their targets may play roles in actin cytoskeleton regulation and vesicle trafficking.
- School of Life Science and Technology, Harbin Institute of Technology, Harbin 150001, China.
Organizational Affiliation: