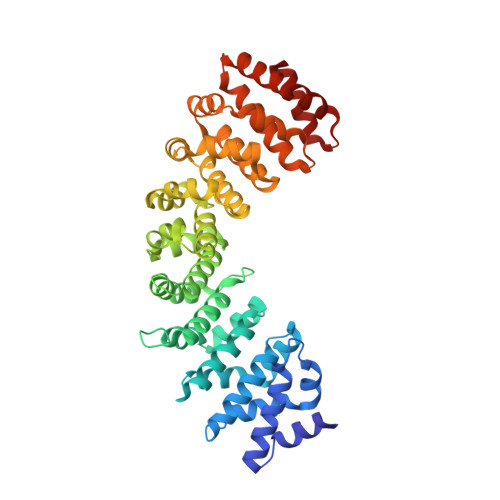
Crystallographic data of an importin-alpha 3 dimer in which the two protomers are bridged by a bipartite nuclear localization signal.
Matsuura, Y.(2023) Data Brief 47: 108988-108988
- PubMed: 36875212
- DOI: https://doi.org/10.1016/j.dib.2023.108988
- Primary Citation of Related Structures:
8HKW - PubMed Abstract:
53BP1 (TP53-binding protein 1), a key player in DNA double-strand break repair, has a classical bipartite nuclear localization signal (NLS) of sequence 1666-GKRKLITSEEERSPAKRGRKS-1686 that binds to importin-α, a nuclear import adaptor protein. Nucleoporin Nup153 is involved in nuclear import of 53BP1, and the binding of Nup153 to importin-α has been proposed to promote efficient import of classical NLS-containing proteins. Here, the ARM-repeat domain of human importin-α3 bound to 53BP1 NLS was crystallized in the presence of a synthetic peptide corresponding to the extreme C-terminus of Nup153 (sequence: 1459-GTSFSGRKIKTAVRRRK-1475). The crystal belonged to space group I 2, with unit-cell parameters a = 95.70, b = 79.60, c = 117.44 Å, β = 95.57°. The crystal diffracted X-rays to 1.9 Å resolution, and the structure was solved by molecular replacement. The asymmetric unit contained two molecules of importin-α3 and two molecules of 53BP1 NLS. Although no convincing density was observed for the Nup153 peptide, the electron density corresponding to 53BP1 NLS was unambiguous and continuous along the entire length of the bipartite NLS. The structure revealed a novel dimer of importin-α3, in which two protomers of importin-α3 are bridged by the bipartite NLS of 53BP1. In this structure, the upstream basic cluster of the NLS is bound to the minor NLS-binding site of one protomer of importin-α3, whereas the downstream basic cluster of the same chain of NLS is bound to the major NLS-binding site of another protomer of importin-α3. This quaternary structure is distinctly different from the previously determined crystal structure of mouse importin-α1 bound to the 53BP1 NLS. The atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession code 8HKW).
- Department of Pharmaceutical Sciences, School of Pharmacy, International University of Health and Welfare, Tochigi 324-8501, Japan.
Organizational Affiliation: