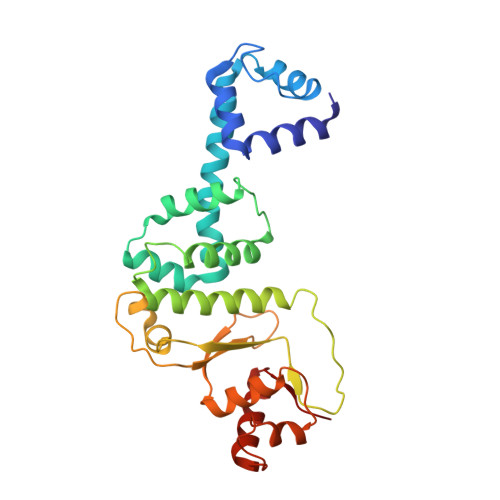
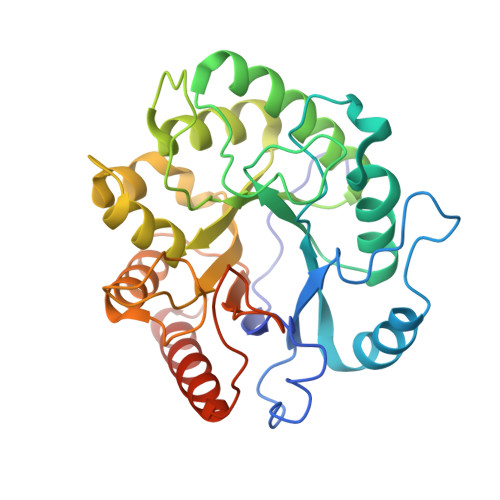
Structures of the holoenzyme TglHI required for 3-thiaglutamate biosynthesis.
Zheng, Y., Xu, X., Fu, X., Zhou, X., Dou, C., Yu, Y., Yan, W., Yang, J., Xiao, M., van der Donk, W.A., Zhu, X., Cheng, W.(2023) Structure 31: 1220-1232.e5
- PubMed: 37652001
- DOI: https://doi.org/10.1016/j.str.2023.08.004
- Primary Citation of Related Structures:
8HCI, 8HI7, 8HI8 - PubMed Abstract:
Structural diverse natural products like ribosomally synthesized and posttranslationally modified peptides (RiPPs) display a wide range of biological activities. Currently, the mechanism of an uncommon reaction step during the biosynthesis of 3-thiaglutamate (3-thiaGlu) is poorly understood. The removal of the β-carbon from the Cys in the TglA-Cys peptide catalyzed by the TglHI holoenzyme remains elusive. Here, we present three crystal structures of TglHI complexes with and without bound iron, which reveal that the catalytic pocket is formed by the interaction of TglH-TglI and that its activation is conformation dependent. Biochemical assays suggest a minimum of two iron ions in the active cluster, and we identify the position of a third iron site. Collectively, our study offers insights into the activation and catalysis mechanisms of the non-heme dioxygen-dependent holoenzyme TglHI. Additionally, it highlights the evolutionary and structural conservation in the DUF692 family of biosynthetic enzymes that produce diverse RiPPs.
- Division of Respiratory and Critical Care Medicine, State Key Laboratory of Biotherapy, West China Hospital of Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu 610041, China.
Organizational Affiliation: