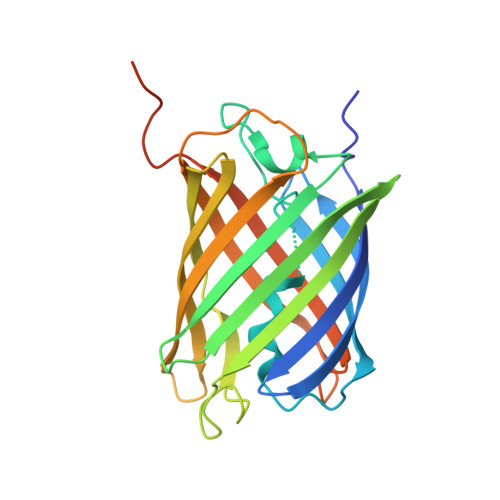
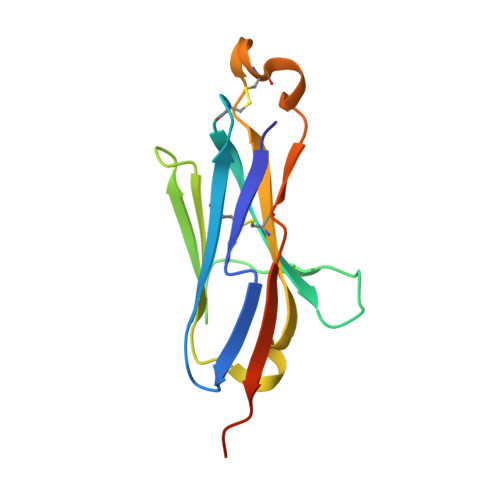
Selection, identification and crystal structure of shark-derived single-domain antibodies against a green fluorescent protein.
Chen, Y.L., Xie, X.X., Zheng, P., Zhu, C., Ma, H., Khalid, Z., Xie, Y.J., Dang, Y.Z., Ye, Y., Sheng, N., Zhong, N., Lei, W.H., Zhang, C., Zhang, L.J., Jin, T., Cao, M.J.(2023) Int J Biol Macromol 247: 125852-125852
- PubMed: 37460076
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.125852
- Primary Citation of Related Structures:
8HGI - PubMed Abstract:
Shark variable domain of new antigen receptors (VNARs) are the smallest naturally occurring binding domains with properties of low complexity, small size, cytoplasmic expression, and ease of engineering. Green fluorescent protein (GFP) molecules have been analyzed in conventional microscopy, but their spectral characteristics preclude their use in techniques offering substantially higher resolution. Besides, the GFP molecules can be quenched in acidic environment, which makes it necessary to develop anti-GFP antibody to solve these problems. In view of the diverse applications of GFP and unique physicochemical features of VNAR, the present study aims to generate VNARs against GFP. Here, we identified 36 VNARs targeting eCGP123, an extremely stable GFP, by phage display from three immunized sharks. These VNARs bound to eCGP123 with affinity constant K D values ranging from 6.76 to 605 nM. Among them, two lead VNARs named aGFP-14 and aGFP-15 with nanomolar eCGP123-binding affinity were selected for in-depth characterization. aGFP-14 and aGFP-15 recognized similar epitopes on eCGP123. X-ray crystallography studies clarified the mechanism by which aGFP14 interacts with eCGP123. aGFP-14 also showed cross-reaction with EGFP, with K D values of 47.2 nM. Finally, immunostaining analyses demonstrated that aGFP-14 was able to bind effectively to the EGFP expressed in both cultured cells and mouse brain tissues, and can be used as a fluorescence amplifier for EGFP. Our research demonstrates a feasible idea for the screening and production of shark-derived VNARs. The two high-affinity VNARs developed in the study contribute to the diversity of GFP sdAbs and may enhance the applications of GFP.
- College of Ocean Food and Biological Engineering, Jimei University, Xiamen 361021, China.
Organizational Affiliation: