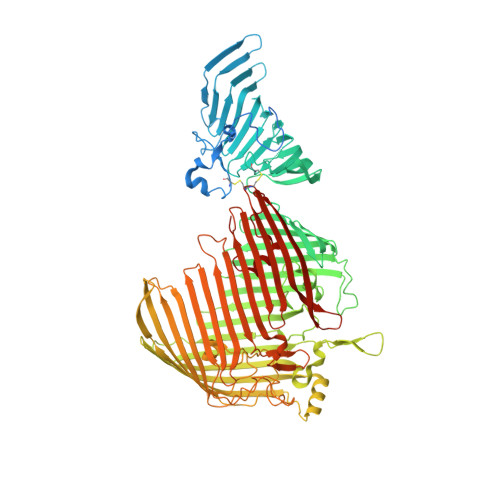
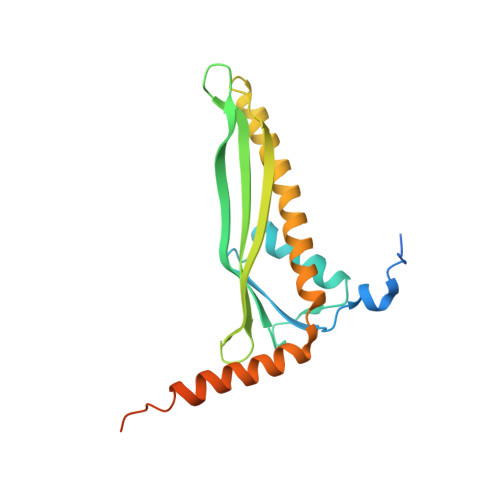
Surface lipoprotein sorting by crosstalk between Lpt and Lol pathways in gram-negative bacteria
Luo, Q., Wang, C., Qiao, S., Yu, S., Chen, L., Kim, S., Wang, K., Zheng, J., Zhang, Y., Wu, F., Lei, X., Lou, J., Hennig, M., Im, W., Miao, L., Zhou, M., Bei, W., Huang, Y.(2025) Nat Commun 16: 4357