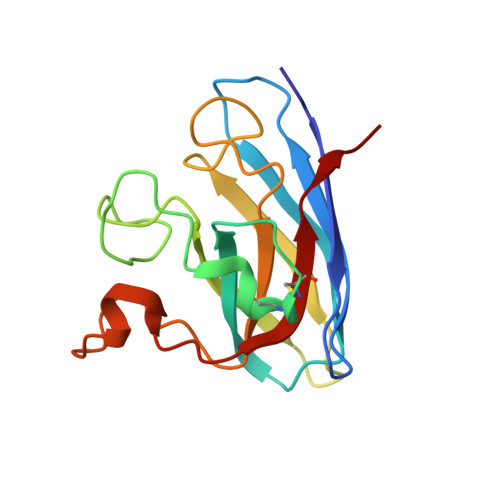
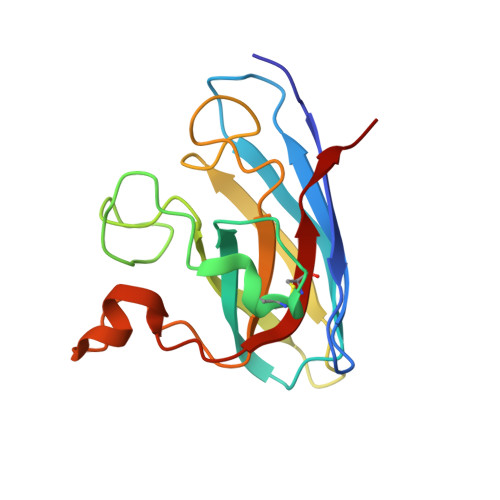
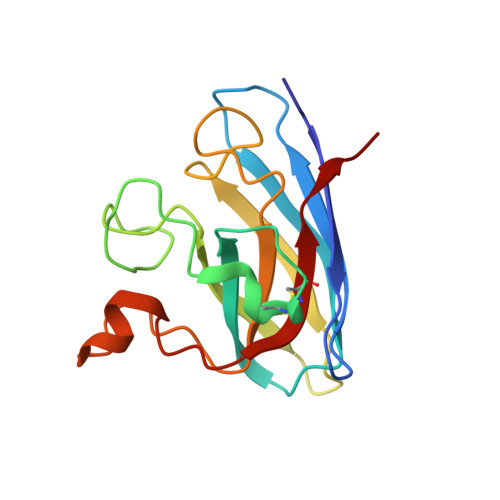
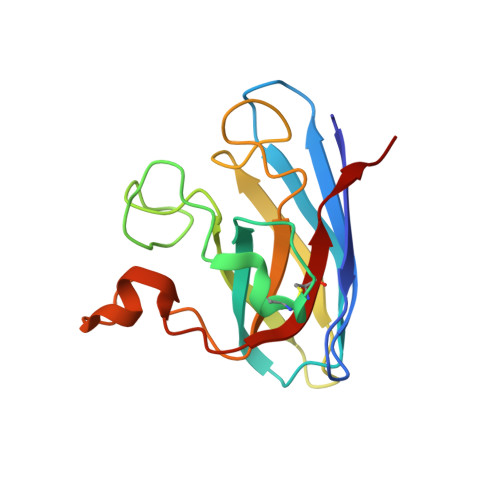
Structure-based discovery of an antipsychotic drug, paliperidone, as a modulator of human superoxide dismutase 1: a potential therapeutic target in amyotrophic lateral sclerosis.
Aouti, S., Padavattan, S., Padmanabhan, B.(2023) Acta Crystallogr D Struct Biol 79: 531-544
- PubMed: 37204819
- DOI: https://doi.org/10.1107/S2059798323003649
- Primary Citation of Related Structures:
8GSQ - PubMed Abstract:
Aggregates of the antioxidant superoxide dismutase 1 (SOD1) are one of the major contributors to the pathogenesis of amyotrophic lateral sclerosis (ALS). Mutations in SOD1 lead to an unstable structure and aggregation that perturbs the balance of reactive oxygen species in cells. Oxidation damage to the solvent-exposed Trp32 also causes aggregation of SOD1. Here, the FDA-approved antipsychotic drug paliperidone is identified to interact with Trp32 of SOD1 by structure-based pharmacophore mapping and crystallographic studies. Paliperidone is used for the treatment of schizophrenia. The crystal structure of the complex with SOD1, refined to 2.1 Å resolution, revealed that the ligand binds to the SOD1 β-barrel in the β-strand 2 and 3 regions, which are known to scaffold SOD1 fibrillation. The drug also makes substantial π-π interaction with Trp32. Microscale thermophoresis studies confirm significant binding affinity of the compound, suggesting that the ligand can inhibit or prevent tryptophan oxidation. Thus, the antipsychotic drug paliperidone or a derivative may avert SOD1 aggregation and can be used as a lead for ALS drug development.
- Department of Biophysics, National Institute of Mental Health and Neurosciences, Bengaluru, Karnataka 560029, India.
Organizational Affiliation: