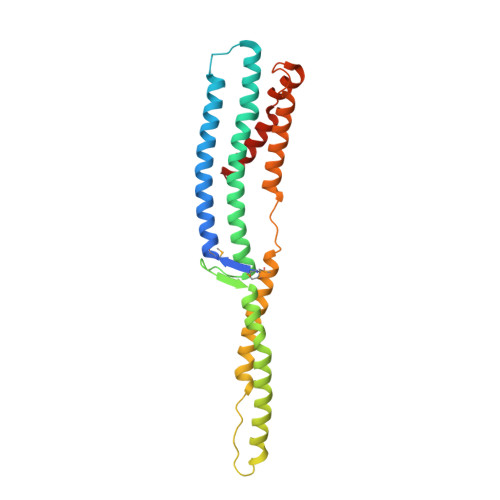
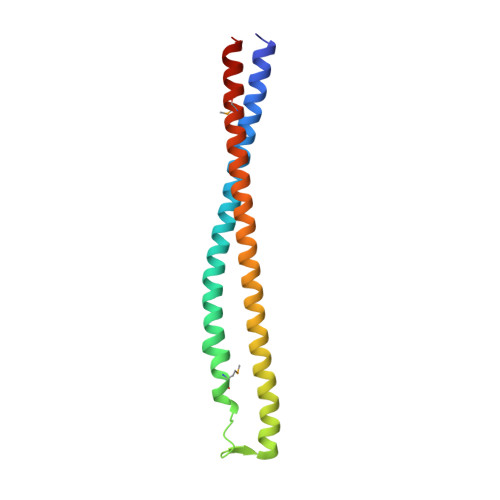
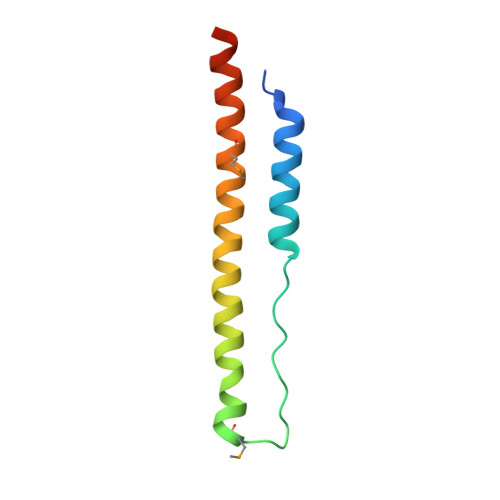
Structure of a tripartite protein complex that targets toxins to the type VII secretion system.
Klein, T.A., Shah, P.Y., Gkragkopoulou, P., Grebenc, D.W., Kim, Y., Whitney, J.C.(2024) Proc Natl Acad Sci U S A 121: e2312455121-e2312455121
- PubMed: 38194450
- DOI: https://doi.org/10.1073/pnas.2312455121
- Primary Citation of Related Structures:
8GMH - PubMed Abstract:
Type VII secretion systems are membrane-embedded nanomachines used by Gram-positive bacteria to export effector proteins from the cytoplasm to the extracellular environment. Many of these effectors are polymorphic toxins comprised of an N-terminal Leu-x-Gly (LXG) domain of unknown function and a C-terminal toxin domain that inhibits the growth of bacterial competitors. In recent work, it was shown that LXG effectors require two cognate Lap proteins for T7SS-dependent export. Here, we present the 2.6 Å structure of the LXG domain of the TelA toxin from the opportunistic pathogen Streptococcus intermedius in complex with both of its cognate Lap targeting factors. The structure reveals an elongated α-helical bundle within which each Lap protein makes extensive hydrophobic contacts with either end of the LXG domain. Remarkably, despite low overall sequence identity, we identify striking structural similarity between our LXG complex and PE-PPE heterodimers exported by the distantly related ESX type VII secretion systems of Mycobacteria implying a conserved mechanism of effector export among diverse Gram-positive bacteria. Overall, our findings demonstrate that LXG domains, in conjunction with their cognate Lap targeting factors, represent a tripartite secretion signal for a widespread family of T7SS toxins.
- Michael DeGroote Institute for Infectious Disease Research, McMaster University, Hamilton, ON L8S 4K1, Canada.
Organizational Affiliation: