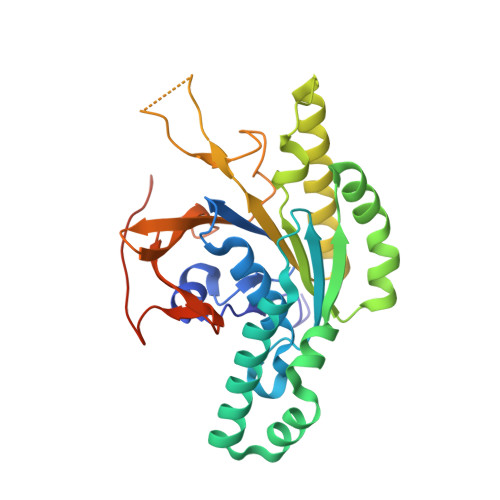
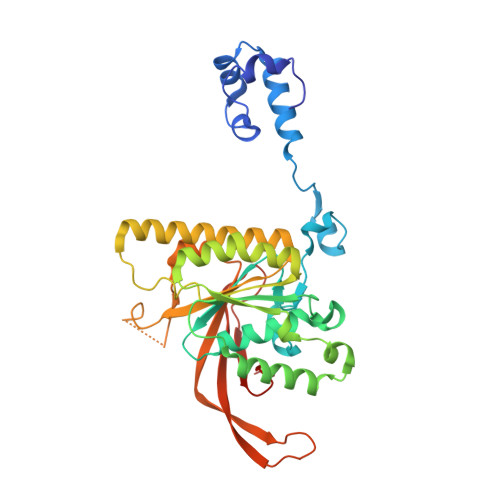
RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Longo, M.A., Roy, S., Chen, Y., Tomaszowski, K.H., Arvai, A.S., Pepper, J.T., Boisvert, R.A., Kunnimalaiyaan, S., Keshvani, C., Schild, D., Bacolla, A., Williams, G.J., Tainer, J.A., Schlacher, K.(2023) Nat Commun 14: 4445-4445
- PubMed: 37488098
- DOI: https://doi.org/10.1038/s41467-023-40096-1
- Primary Citation of Related Structures:
8GJ8, 8GJ9, 8GJA - PubMed Abstract:
RAD51C is an enigmatic predisposition gene for breast, ovarian, and prostate cancer. Currently, missing structural and related functional understanding limits patient mutation interpretation to homology-directed repair (HDR) function analysis. Here we report the RAD51C-XRCC3 (CX3) X-ray co-crystal structure with bound ATP analog and define separable RAD51C replication stability roles informed by its three-dimensional structure, assembly, and unappreciated polymerization motif. Mapping of cancer patient mutations as a functional guide confirms ATP-binding matching RAD51 recombinase, yet highlights distinct CX3 interfaces. Analyses of CRISPR/Cas9-edited human cells with RAD51C mutations combined with single-molecule, single-cell and biophysics measurements uncover discrete CX3 regions for DNA replication fork protection, restart and reversal, accomplished by separable functions in DNA binding and implied 5' RAD51 filament capping. Collective findings establish CX3 as a cancer-relevant replication stress response complex, show how HDR-proficient variants could contribute to tumor development, and identify regions to aid functional testing and classification of cancer mutations.
- Department of Molecular & Cellular Oncology, UT MD Anderson Cancer Center, Houston, TX, USA.
Organizational Affiliation: