Molecular insights into recognition of GUCY2C by T-cell engaging bispecific antibody anti-GUCY2CxCD3.
Rampuria, P., Mosyak, L., Root, A.R., Svenson, K., Agostino, M.J., LaVallie, E.R.(2023) Sci Rep 13: 13408-13408
- PubMed: 37591971
- DOI: https://doi.org/10.1038/s41598-023-40467-0
- Primary Citation of Related Structures:
8GHO, 8GHP - PubMed Abstract:
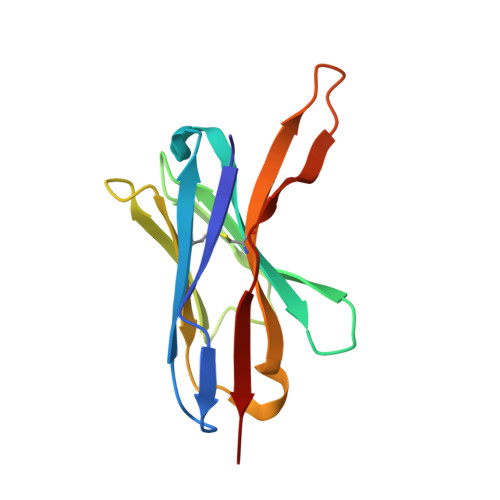
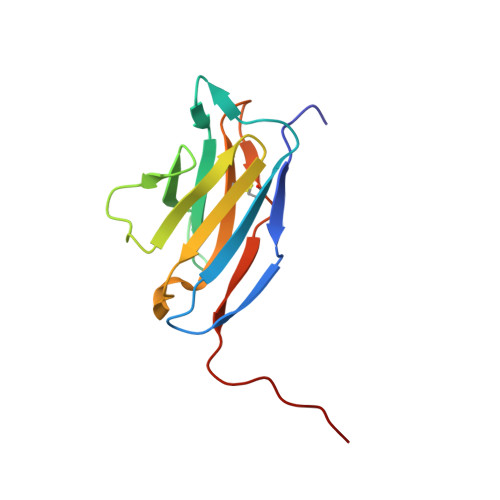
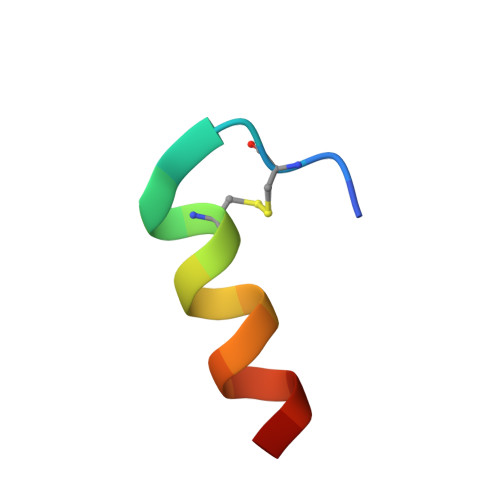
The intestinal epithelial receptor Guanylyl Cyclase C (GUCY2C) is a tumor-associated cell surface antigen expressed across gastrointestinal malignancies that can serve as an efficacious target for colorectal cancer immunotherapy. Here, we describe a yeast surface-display approach combined with an orthogonal peptide-based mapping strategy to identify the GUCY2C binding epitope of a novel anti-GUCY2CxCD3 bispecific antibody (BsAb) that recently advanced into the clinic for the treatment of cancer. The target epitope was localized to the N-terminal helix H2 of human GUCY2C, which enabled the determination of the crystal structure of the minimal GUCY2C epitope in complex with the anti-GUCY2C antibody domain. To understand if this minimal epitope covers the entire antibody binding region and to investigate the impact of epitope position on the antibody's activity, we further determined the structure of this interaction in the context of the full-length extracellular domain (ECD) of GUCY2C. We found that this epitope is positioned on the protruding membrane-distal helical region of GUCY2C and that its specific location on the surface of GUCY2C dictates the close spatial proximity of the two antigen arms in a diabody arrangement essential to the tumor killing activity of GUCY2CxCD3 BsAb.
- Biomedicine Design, Pfizer Inc., 610 Main St., Cambridge, MA, 02139, USA. pragya.rampuria@pfizer.com.
Organizational Affiliation: