Implementing Logic Gates by DNA Crystal Engineering.
Zhang, C., Paluzzi, V.E., Sha, R., Jonoska, N., Mao, C.(2023) Adv Mater 35: e2302345-e2302345
- PubMed: 37220213
- DOI: https://doi.org/10.1002/adma.202302345
- Primary Citation of Related Structures:
8GH5 - PubMed Abstract:
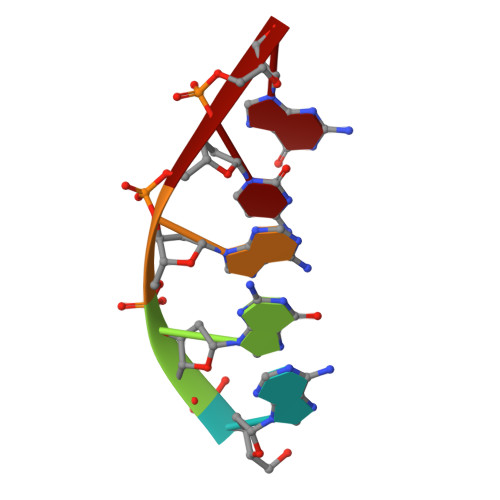
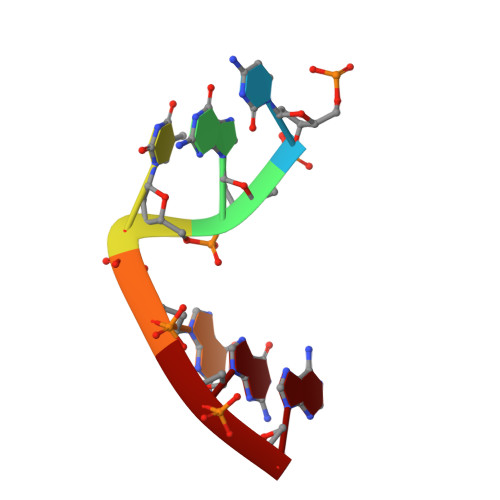
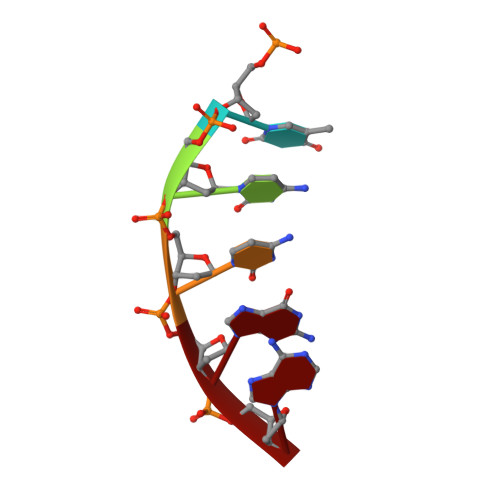
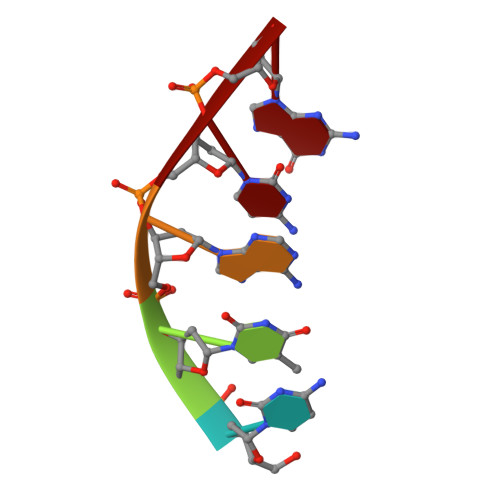
DNA self-assembly computation is attractive for its potential to perform massively parallel information processing at the molecular level while at the same time maintaining its natural biocompatibility. It has been extensively studied at the individual molecule level, but not as much as ensembles in 3D. Here, the feasibility of implementing logic gates, the basic computation operations, in large ensembles: macroscopic, engineered 3D DNA crystals is demonstrated. The building blocks are the recently developed DNA double crossover-like (DXL) motifs. They can associate with each other via sticky-end cohesion. Common logic gates are realized by encoding the inputs within the sticky ends of the motifs. The outputs are demonstrated through the formation of macroscopic crystals that can be easily observed. This study points to a new direction of construction of complex 3D crystal architectures and DNA-based biosensors with easy readouts.
- Department of Chemistry, Purdue University, West Lafayette, IN, 47907, USA.
Organizational Affiliation: