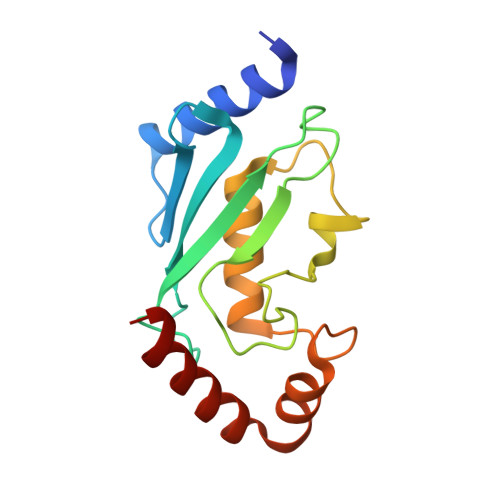
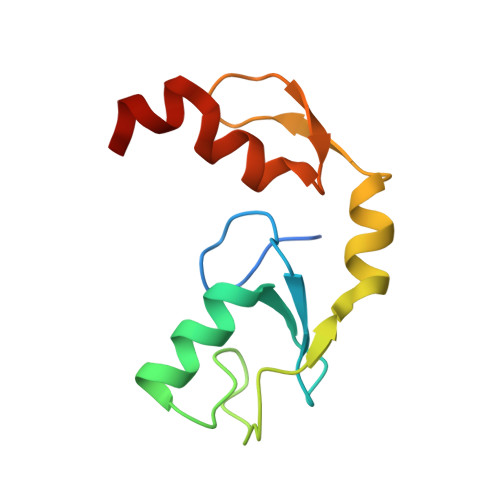
Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Middleton, A.J., Barzak, F.M., Fokkens, T.J., Nguyen, K., Day, C.L.(2023) Structure 31: 1208-1219.e5
- PubMed: 37541247
- DOI: https://doi.org/10.1016/j.str.2023.07.007
- Primary Citation of Related Structures:
8GBQ, 8GCB - PubMed Abstract:
Inflammation is essential for healthy immune function, wound healing, and resolution of infection. RIG-I is a key RNA sensor that initiates an immune response, with activation and termination of RIG-I signaling reliant on its modification with ubiquitin. The RING E3 ubiquitin ligase, RNF125, has a critical role in the attenuation of RIG-I signaling, yet it is not known how RNF125 promotes ubiquitin transfer or how its activity is regulated. Here we show that the E3 ligase activity of RNF125 relies on the first zinc finger (ZF1) as well as the RING domain. Surprisingly, ZF1 helps recruit the E2, while residues N-terminal to the RING domain appear to activate the E2∼Ub conjugate. These discoveries help explain how RNF125 brings about the termination of RIG-I dependent inflammatory responses, and help account for the contribution of RNF125 to disease. This study also reveals a new role for ZF domains in E3 ligases.
- Department of Biochemistry, School of Biomedical Sciences, University of Otago, Dunedin 9054, New Zealand.
Organizational Affiliation: