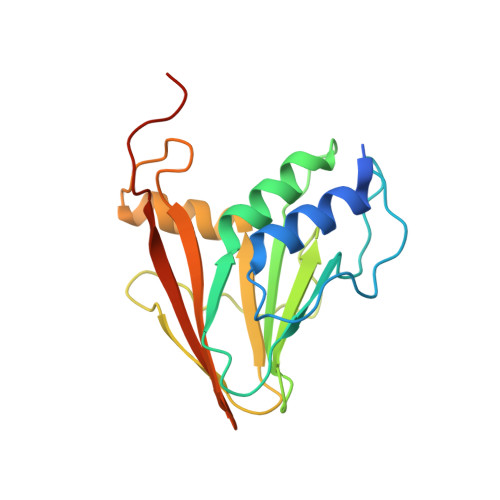
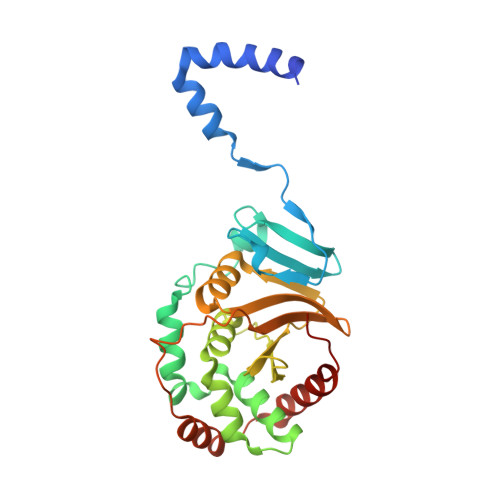
The universal suppressor mutation restores membrane budding defects in the HSV-1 nuclear egress complex by stabilizing the oligomeric lattice.
Draganova, E.B., Wang, H., Wu, M., Liao, S., Vu, A., Gonzalez-Del Pino, G.L., Zhou, Z.H., Roller, R.J., Heldwein, E.E.(2024) PLoS Pathog 20: e1011936-e1011936
- PubMed: 38227586
- DOI: https://doi.org/10.1371/journal.ppat.1011936
- Primary Citation of Related Structures:
8G6D - PubMed Abstract:
Nuclear egress is an essential process in herpesvirus replication whereby nascent capsids translocate from the nucleus to the cytoplasm. This initial step of nuclear egress-budding at the inner nuclear membrane-is coordinated by the nuclear egress complex (NEC). Composed of the viral proteins UL31 and UL34, NEC deforms the membrane around the capsid as the latter buds into the perinuclear space. NEC oligomerization into a hexagonal membrane-bound lattice is essential for budding because NEC mutants designed to perturb lattice interfaces reduce its budding ability. Previously, we identified an NEC suppressor mutation capable of restoring budding to a mutant with a weakened hexagonal lattice. Using an established in-vitro budding assay and HSV-1 infected cell experiments, we show that the suppressor mutation can restore budding to a broad range of budding-deficient NEC mutants thereby acting as a universal suppressor. Cryogenic electron tomography of the suppressor NEC mutant lattice revealed a hexagonal lattice reminiscent of wild-type NEC lattice instead of an alternative lattice. Further investigation using x-ray crystallography showed that the suppressor mutation promoted the formation of new contacts between the NEC hexamers that, ostensibly, stabilized the hexagonal lattice. This stabilization strategy is powerful enough to override the otherwise deleterious effects of mutations that destabilize the NEC lattice by different mechanisms, resulting in a functional NEC hexagonal lattice and restoration of membrane budding.
- Department of Molecular Biology and Microbiology, Tufts University School of Medicine, Boston, Massachusetts, United States of America.
Organizational Affiliation: