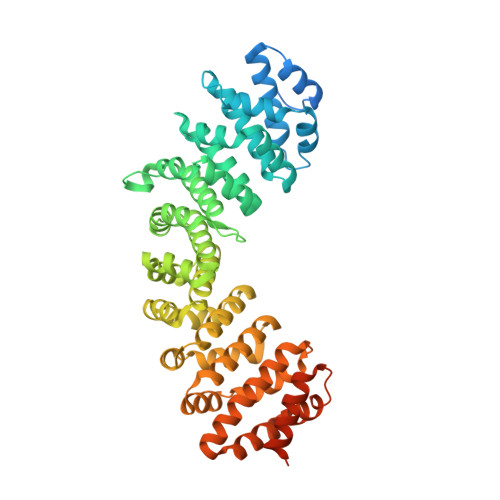
Structural Characterization of Porcine Adeno-Associated Virus Capsid Protein with Nuclear Trafficking Protein Importin Alpha Reveals a Bipartite Nuclear Localization Signal.
Hoad, M., Cross, E.M., Donnelly, C.M., Sarker, S., Roby, J.A., Forwood, J.K.(2023) Viruses 15
- PubMed: 36851528
- DOI: https://doi.org/10.3390/v15020315
- Primary Citation of Related Structures:
8FK3 - PubMed Abstract:
Adeno-associated viruses (AAV) are important vectors for gene therapy, and accordingly, many aspects of their cell transduction pathway have been well characterized. However, the specific mechanisms that AAV virions use to enter the host nucleus remain largely unresolved. We therefore aimed to reveal the interactions between the AAV Cap protein and the nuclear transport protein importin alpha (IMPα) at an atomic resolution. Herein we expanded upon our earlier research into the Cap nuclear localization signal (NLS) of a porcine AAV isolate, by examining the influence of upstream basic regions (BRs) towards IMPα binding. Using a high-resolution crystal structure, we identified that the IMPα binding determinants of the porcine AAV Cap comprise a bipartite NLS with an N-terminal BR binding at the minor site of IMPα, and the previously identified NLS motif binding at the major site. Quantitative assays showed a vast difference in binding affinity between the previously determined monopartite NLS, and bipartite NLS described in this study. Our results provide a detailed molecular view of the interaction between AAV capsids and the nuclear import receptor, and support the findings that AAV capsids enter the nucleus by binding the nuclear import adapter IMPα using the classical nuclear localization pathway.
- School of Dentistry and Medical Sciences, Charles Sturt University, Wagga Wagga, NSW 2678, Australia.
Organizational Affiliation: