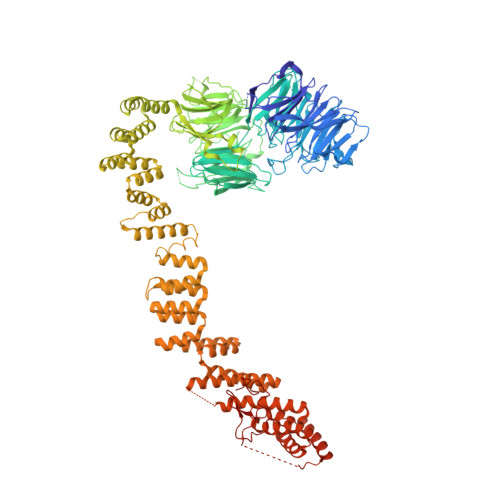
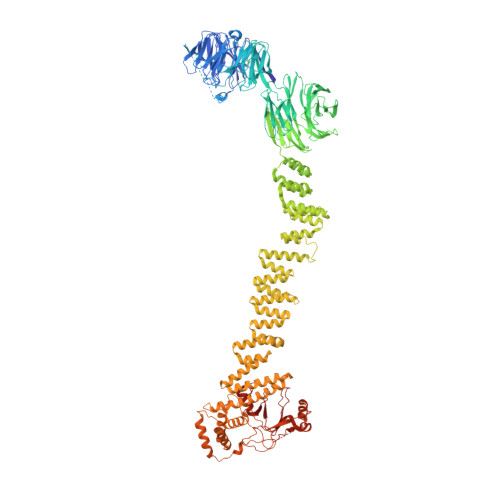
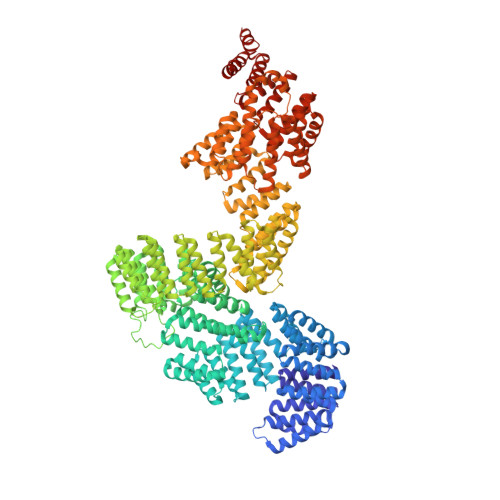
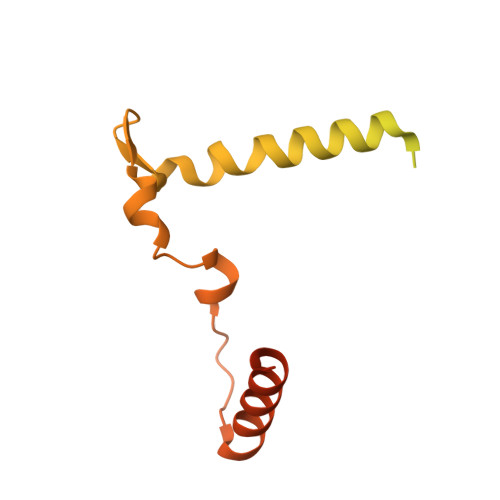
Human IFT-A complex structures provide molecular insights into ciliary transport.
Jiang, M., Palicharla, V.R., Miller, D., Hwang, S.H., Zhu, H., Hixson, P., Mukhopadhyay, S., Sun, J.(2023) Cell Res 33: 288-298
- PubMed: 36775821
- DOI: https://doi.org/10.1038/s41422-023-00778-3
- Primary Citation of Related Structures:
8FGW, 8FH3 - PubMed Abstract:
Intraflagellar transport (IFT) complexes, IFT-A and IFT-B, form bidirectional trains that move along the axonemal microtubules and are essential for assembling and maintaining cilia. Mutations in IFT subunits lead to numerous ciliopathies involving multiple tissues. However, how IFT complexes assemble and mediate cargo transport lacks mechanistic understanding due to missing high-resolution structural information of the holo-complexes. Here we report cryo-EM structures of human IFT-A complexes in the presence and absence of TULP3 at overall resolutions of 3.0-3.9 Å. IFT-A adopts a "lariat" shape with interconnected core and peripheral subunits linked by structurally vital zinc-binding domains. TULP3, the cargo adapter, interacts with IFT-A through its N-terminal region, and interface mutations disrupt cargo transport. We also determine the molecular impacts of disease mutations on complex formation and ciliary transport. Our work reveals IFT-A architecture, sheds light on ciliary transport and IFT train formation, and enables the rationalization of disease mutations in ciliopathies.
- Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN, USA.
Organizational Affiliation: