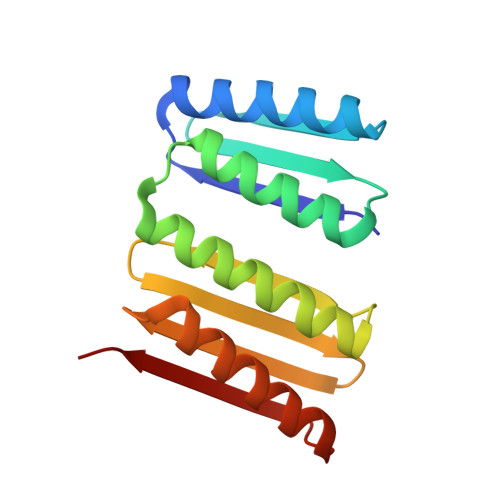
Design of amyloidogenic peptide traps.
Sahtoe, D.D., Andrzejewska, E.A., Han, H.L., Rennella, E., Schneider, M.M., Meisl, G., Ahlrichs, M., Decarreau, J., Nguyen, H., Kang, A., Levine, P., Lamb, M., Li, X., Bera, A.K., Kay, L.E., Knowles, T.P.J., Baker, D.(2024) Nat Chem Biol 20: 981-990
- PubMed: 38503834
- DOI: https://doi.org/10.1038/s41589-024-01578-5
- Primary Citation of Related Structures:
8FG6 - PubMed Abstract:
Segments of proteins with high β-strand propensity can self-associate to form amyloid fibrils implicated in many diseases. We describe a general approach to bind such segments in β-strand and β-hairpin conformations using de novo designed scaffolds that contain deep peptide-binding clefts. The designs bind their cognate peptides in vitro with nanomolar affinities. The crystal structure of a designed protein-peptide complex is close to the design model, and NMR characterization reveals how the peptide-binding cleft is protected in the apo state. We use the approach to design binders to the amyloid-forming proteins transthyretin, tau, serum amyloid A1 and amyloid β 1-42 (Aβ42). The Aβ binders block the assembly of Aβ fibrils as effectively as the most potent of the clinically tested antibodies to date and protect cells from toxic Aβ42 species.
- Department of Biochemistry, University of Washington, Seattle, WA, USA. d.sahtoe@hubrecht.eu.
Organizational Affiliation: