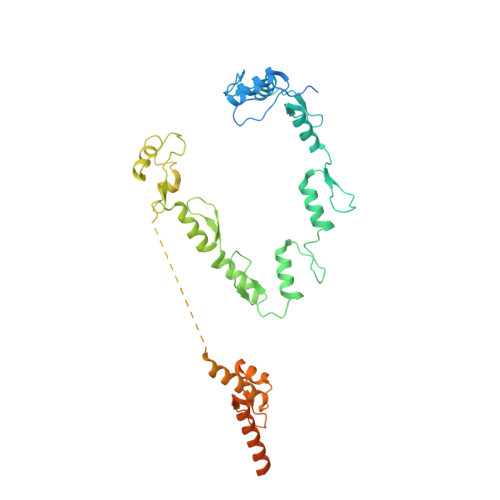
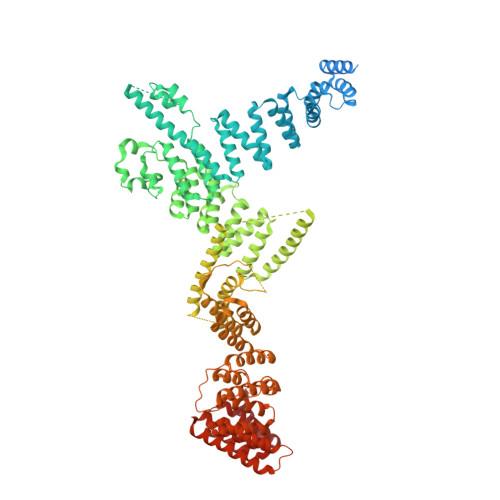
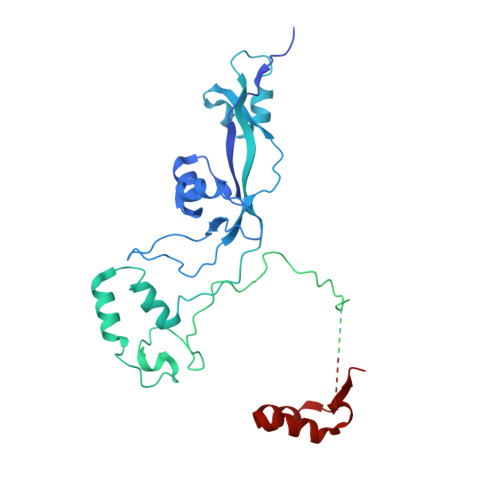
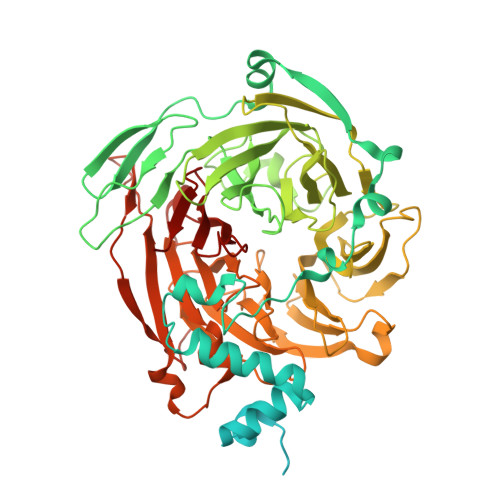
Structural basis of TFIIIC-dependent RNA polymerase III transcription initiation.
Talyzina, A., Han, Y., Banerjee, C., Fishbain, S., Reyes, A., Vafabakhsh, R., He, Y.(2023) Mol Cell 83: 2641
- PubMed: 37402369
- DOI: https://doi.org/10.1016/j.molcel.2023.06.015
- Primary Citation of Related Structures:
8FFZ - PubMed Abstract:
RNA polymerase III (Pol III) is responsible for transcribing 5S ribosomal RNA (5S rRNA), tRNAs, and other short non-coding RNAs. Its recruitment to the 5S rRNA promoter requires transcription factors TFIIIA, TFIIIC, and TFIIIB. Here, we use cryoelectron microscopy (cryo-EM) to visualize the S. cerevisiae complex of TFIIIA and TFIIIC bound to the promoter. Gene-specific factor TFIIIA interacts with DNA and acts as an adaptor for TFIIIC-promoter interactions. We also visualize DNA binding of TFIIIB subunits, Brf1 and TBP (TATA-box binding protein), which results in the full-length 5S rRNA gene wrapping around the complex. Our smFRET study reveals that the DNA within the complex undergoes both sharp bending and partial dissociation on a slow timescale, consistent with the model predicted from our cryo-EM results. Our findings provide new insights into the transcription initiation complex assembly on the 5S rRNA promoter and allow us to directly compare Pol III and Pol II transcription adaptations.
- Department of Molecular Biosciences, Northwestern University, Evanston, IL, USA; Interdisciplinary Biological Sciences Program, Northwestern University, Evanston, IL, USA.
Organizational Affiliation: