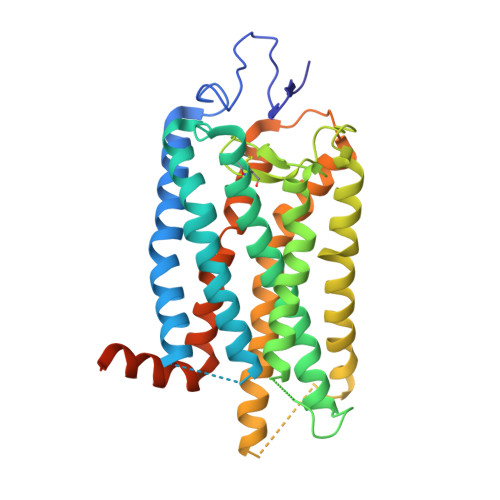
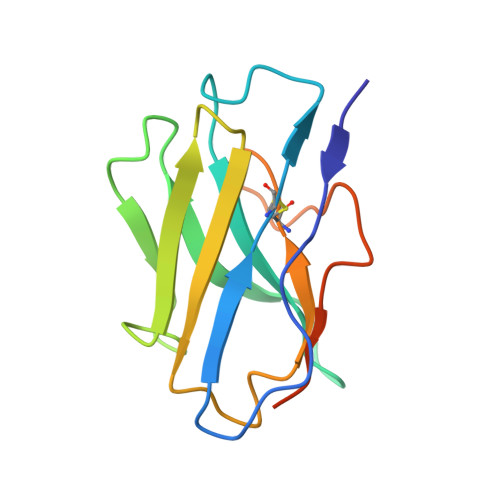
Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Wu, A., Salom, D., Hong, J.D., Tworak, A., Watanabe, K., Pardon, E., Steyaert, J., Kandori, H., Katayama, K., Kiser, P.D., Palczewski, K.(2023) Nat Commun 14: 5209-5209
- PubMed: 37626045
- DOI: https://doi.org/10.1038/s41467-023-40911-9
- Primary Citation of Related Structures:
8FCZ, 8FD0, 8FD1 - PubMed Abstract:
Rhodopsin is a prototypical G protein-coupled receptor (GPCR) critical for vertebrate vision. Research on GPCR signaling states has been facilitated using llama-derived nanobodies (Nbs), some of which bind to the intracellular surface to allosterically modulate the receptor. Extracellularly binding allosteric nanobodies have also been investigated, but the structural basis for their activity has not been resolved to date. Here, we report a library of Nbs that bind to the extracellular surface of rhodopsin and allosterically modulate the thermodynamics of its activation process. Crystal structures of Nb2 in complex with native rhodopsin reveal a mechanism of allosteric modulation involving extracellular loop 2 and native glycans. Nb2 binding suppresses Schiff base deprotonation and hydrolysis and prevents intracellular outward movement of helices five and six - a universal activation event for GPCRs. Nb2 also mitigates protein misfolding in a disease-associated mutant rhodopsin. Our data show the power of nanobodies to modulate the photoactivation of rhodopsin and potentially serve as therapeutic agents for disease-associated rhodopsin misfolding.
- Department of Ophthalmology, Gavin Herbert Eye Institute, University of California, Irvine, CA, 92697, USA.
Organizational Affiliation: