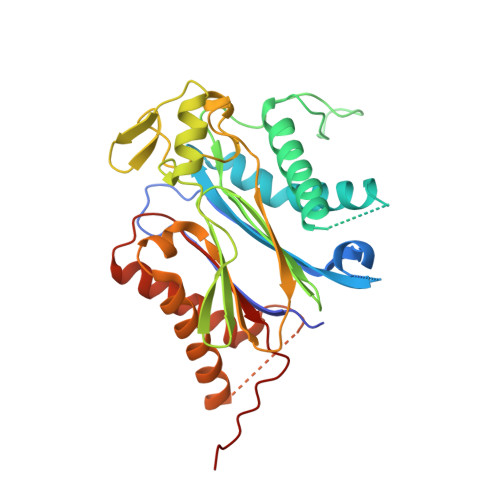
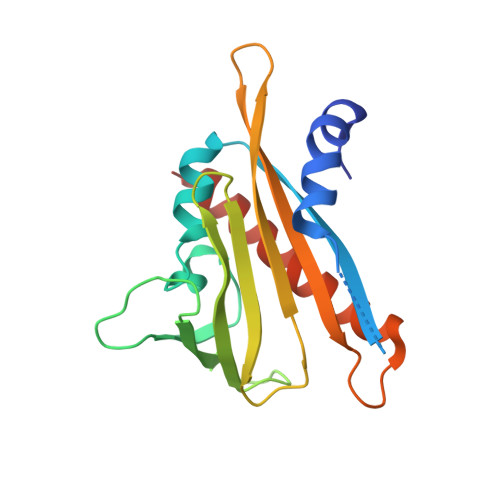
An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity.
Park, S.Y., Qiu, J., Wei, S., Peterson, F.C., Beltran, J., Medina-Cucurella, A.V., Vaidya, A.S., Xing, Z., Volkman, B.F., Nusinow, D.A., Whitehead, T.A., Wheeldon, I., Cutler, S.R.(2024) Nat Chem Biol 20: 103-110
- PubMed: 37872402
- DOI: https://doi.org/10.1038/s41589-023-01447-7
- Primary Citation of Related Structures:
8EY0 - PubMed Abstract:
Plants sense abscisic acid (ABA) using chemical-induced dimerization (CID) modules, including the receptor PYR1 and HAB1, a phosphatase inhibited by ligand-activated PYR1. This system is unique because of the relative ease with which ligand recognition can be reprogrammed. To expand the PYR1 system, we designed an orthogonal '*' module, which harbors a dimer interface salt bridge; X-ray crystallographic, biochemical and in vivo analyses confirm its orthogonality. We used this module to create PYR1* MANDI /HAB1* and PYR1* AZIN /HAB1*, which possess nanomolar sensitivities to their activating ligands mandipropamid and azinphos-ethyl. Experiments in Arabidopsis thaliana and Saccharomyces cerevisiae demonstrate the sensitive detection of banned organophosphate contaminants using living biosensors and the construction of multi-input/output genetic circuits. Our new modules enable ligand-programmable multi-channel CID systems for plant and eukaryotic synthetic biology that can empower new plant-based and microbe-based sensing modalities.
- Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA, USA.
Organizational Affiliation: