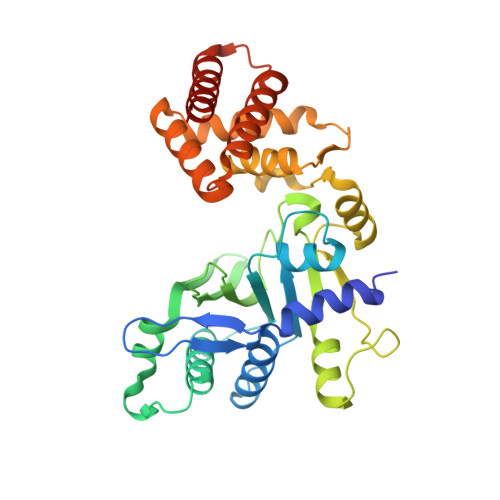
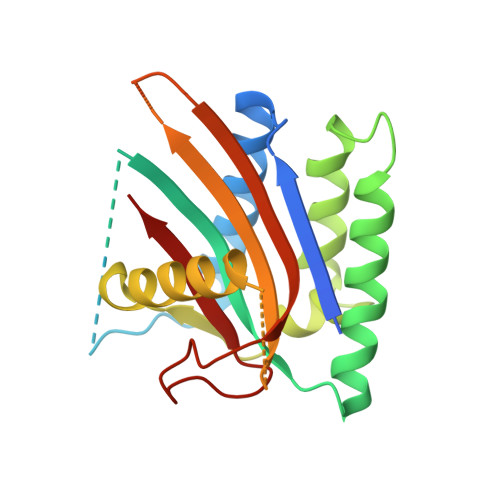
Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Liu, S., Chen, H., Yin, Y., Lu, D., Gao, G., Li, J., Bai, X.C., Zhang, X.(2023) Sci Adv 9: eadf5583-eadf5583
- PubMed: 37018411
- DOI: https://doi.org/10.1126/sciadv.adf5583
- Primary Citation of Related Structures:
8EQB, 8EXE, 8EXF - PubMed Abstract:
The FAM46 (also known as TENT5) proteins are noncanonical poly(A) polymerases (PAPs) implicated in regulating RNA stability. The regulatory mechanisms of FAM46 are poorly understood. Here, we report that the nuclear protein BCCIPα, but not the alternatively spliced isoform BCCIPβ, binds FAM46 and inhibits their PAP activity. Unexpectedly, our structures of the FAM46A/BCCIPα and FAM46C/BCCIPα complexes show that, despite sharing most of the sequence and differing only at the C-terminal portion, BCCIPα adopts a unique structure completely different from BCCIPβ. The distinct C-terminal segment of BCCIPα supports the adoption of the unique fold but does not directly interact with FAM46. The β sheets in BCCIPα and FAM46 pack side by side to form an extended β sheet. A helix-loop-helix segment in BCCIPα inserts into the active site cleft of FAM46, thereby inhibiting the PAP activity. Our results together show that the unique fold of BCCIPα underlies its interaction with and functional regulation of FAM46.
- Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: