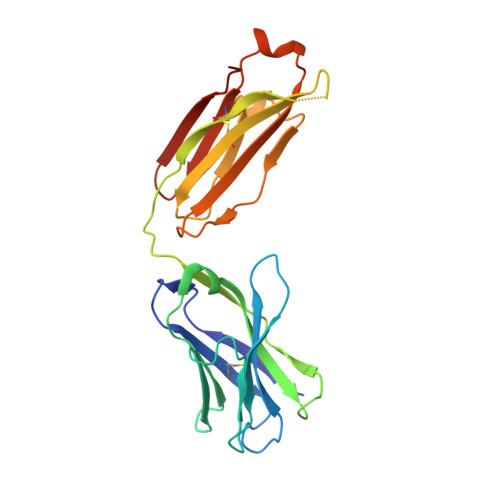
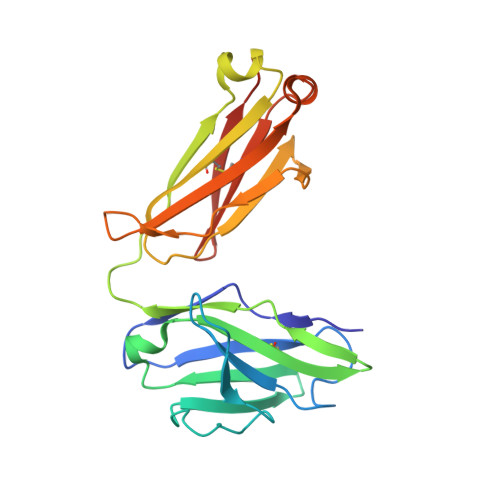
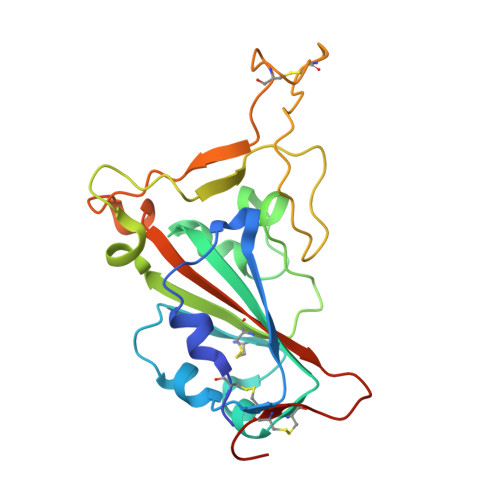
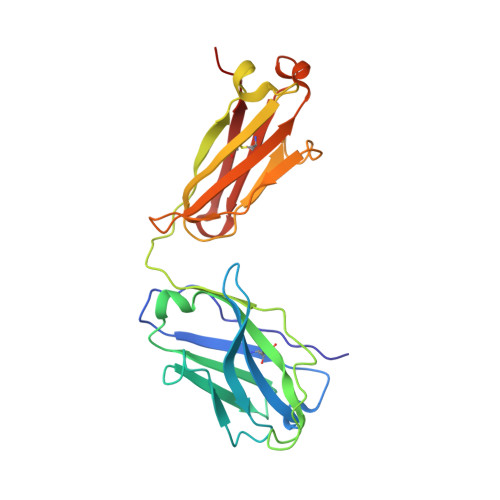
Targeting the Spike Receptor Binding Domain Class V Cryptic Epitope by an Antibody with Pan-Sarbecovirus Activity.
Jensen, J.L., Sankhala, R.S., Dussupt, V., Bai, H., Hajduczki, A., Lal, K.G., Chang, W.C., Martinez, E.J., Peterson, C.E., Golub, E.S., Rees, P.A., Mendez-Rivera, L., Zemil, M., Kavusak, E., Mayer, S.V., Wieczorek, L., Kannan, S., Doranz, B.J., Davidson, E., Yang, E.S., Zhang, Y., Chen, M., Choe, M., Wang, L., Gromowski, G.D., Koup, R.A., Michael, N.L., Polonis, V.R., Rolland, M., Modjarrad, K., Krebs, S.J., Joyce, M.G.(2023) J Virol 97: e0159622-e0159622
- PubMed: 37395646
- DOI: https://doi.org/10.1128/jvi.01596-22
- Primary Citation of Related Structures:
8EOO - PubMed Abstract:
Novel therapeutic monoclonal antibodies (MAbs) must accommodate comprehensive breadth of activity against diverse sarbecoviruses and high neutralization potency to overcome emerging variants. Here, we report the crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor binding domain (RBD) in complex with MAb WRAIR-2063, a moderate-potency neutralizing antibody with exceptional sarbecovirus breadth, that targets the highly conserved cryptic class V epitope. This epitope overlaps substantially with the spike protein N-terminal domain (NTD) -interacting region and is exposed only when the spike is in the open conformation, with one or more RBDs accessible. WRAIR-2063 binds the RBD of SARS-CoV-2 WA-1, all variants of concern (VoCs), and clade 1 to 4 sarbecoviruses with high affinity, demonstrating the conservation of this epitope and potential resiliency against variation. We compare structural features of additional class V antibodies with their reported neutralization capacity to further explore the utility of the class V epitope as a pan-sarbecovirus vaccine and therapeutic target. IMPORTANCE Characterization of MAbs against SARS-CoV-2, elicited through vaccination or natural infection, has provided vital immunotherapeutic options for curbing the COVID-19 pandemic and has supplied critical insights into SARS-CoV-2 escape, transmissibility, and mechanisms of viral inactivation. Neutralizing MAbs that target the RBD but do not block ACE2 binding are of particular interest because the epitopes are well conserved within sarbecoviruses and MAbs targeting this area demonstrate cross-reactivity. The class V RBD-targeted MAbs localize to an invariant site of vulnerability, provide a range of neutralization potency, and exhibit considerable breadth against divergent sarbecoviruses, with implications for vaccine and therapeutic development.
- Emerging Infectious Diseases Branch, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Organizational Affiliation: