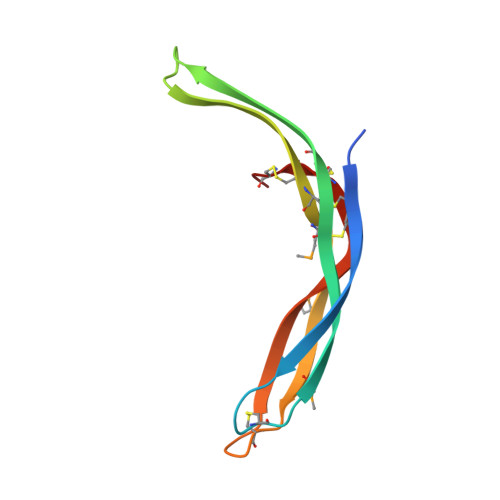
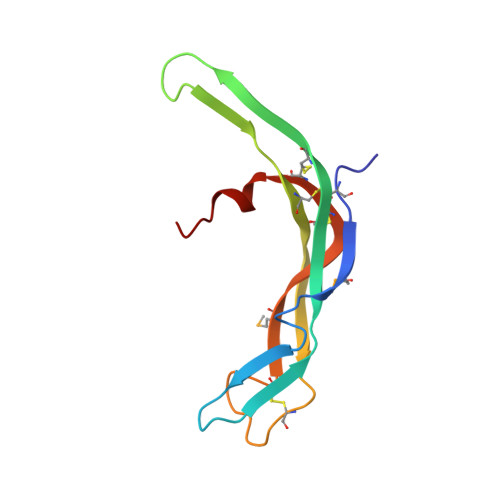
Crystal structure of LGR ligand alpha2/beta5 from Caenorhabditis elegans with implications for the evolution of glycoprotein hormones
Gong, Z., Wang, W., El Omari, K., Lebedev, A.A., Clarke, O.B., Hendrickson, W.A.(2023) Proc Natl Acad Sci U S A 120: e2218630120
- PubMed: 36574673
- DOI: https://doi.org/10.1073/pnas.2218630120
- Primary Citation of Related Structures:
8ENB, 8END, 8ENF - PubMed Abstract:
A family of leucine-rich-repeat-containing G-protein-coupled receptors (LGRs) mediate diverse physiological responses when complexed with their cognate ligands. LGRs are present in all metazoan animals. In humans, the LGR ligands include glycoprotein hormones (GPHs) chorionic gonadotropin (hCG), luteinizing hormone, follicle-stimulating hormone (hFSH), and thyroid-stimulating hormone (hTSH). These hormones are αβ heterodimers of cystine-knot protein chains. LGRs and their ligand chains have coevolved. Ancestral hormone homologs, present in both bilaterian animals and chordates, are identified as α2β5. We have used single-wavelength anomalous diffraction and molecular replacement to determine structures of the α2β5 hormone from Caenorhabditis elegans ( Ce α2β5). Ce α2β5 is unglycosylated, as are many other α2β5 hormones. Both Hs α2β5, the human homolog of Ce α2β5, and hTSH activate the same receptor (hTSHR). Despite having little sequence similarity to vertebrate GPHs, apart from the cysteine patterns from core disulfide bridges, Ce α2β5 is generally similar in structure to these counterparts; however, its α2 and β5 subunits are more symmetric as compared with α and β of hCG and hFSH. This quasisymmetry suggests a hypothetical homodimeric antecedent of the α2β5 and αβ heterodimers. Known structures together with AlphaFold models from the sequences for other LGR ligands provide representatives for the molecular evolution of LGR ligands from early metazoans through the present-day GPHs. The experimental Ce α2β5 structure validates its AlphaFold model, and thus also that for Hs α2β5; and interfacial characteristics in a model for the Hs α2β5:hTSHR complex are similar to those found in an experimental hTSH:hTSHR structure.
- Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032.
Organizational Affiliation: