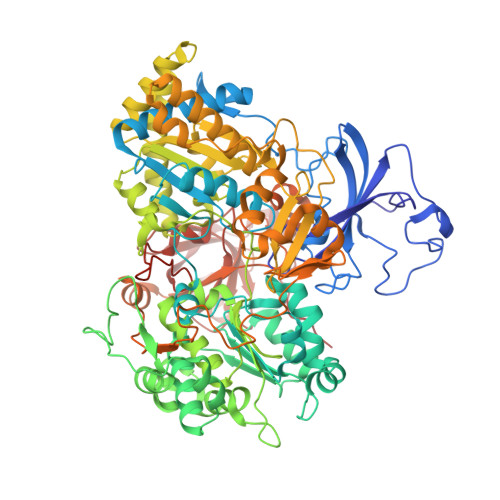
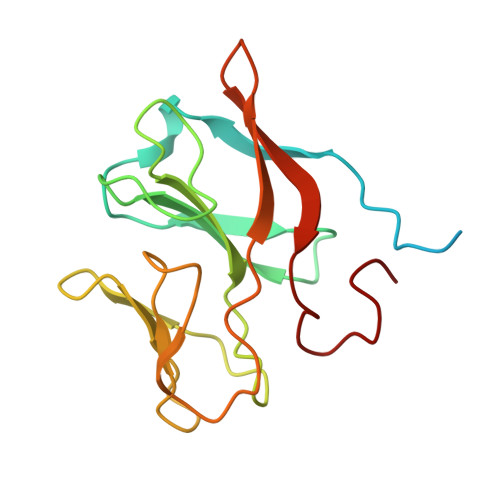
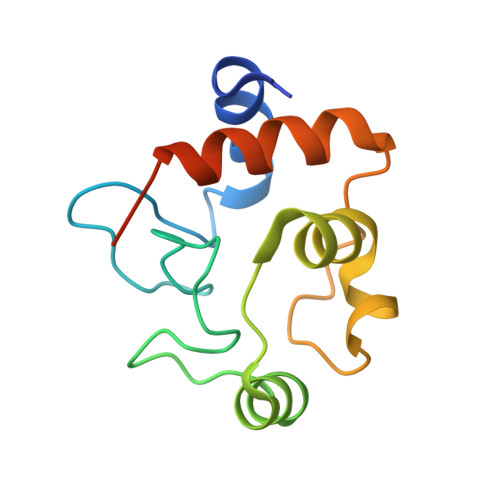
The structure of the complex between the arsenite oxidase from Pseudorhizobium banfieldiae sp. strain NT-26 and its native electron acceptor cytochrome c 552.
Poddar, N., Santini, J.M., Maher, M.J.(2023) Acta Crystallogr D Struct Biol 79: 345-352
- PubMed: 36995233
- DOI: https://doi.org/10.1107/S2059798323002103
- Primary Citation of Related Structures:
8ED4 - PubMed Abstract:
The arsenite oxidase (AioAB) from Pseudorhizobium banfieldiae sp. strain NT-26 catalyzes the oxidation of arsenite to arsenate and transfers electrons to its cognate electron acceptor cytochrome c 552 (cytc 552 ). This activity underpins the ability of this organism to respire using arsenite present in contaminated environments. The crystal structure of the AioAB/cytc 552 electron transfer complex reveals two A 2 B 2 /(cytc 552 ) 2 assemblies per asymmetric unit. Three of the four cytc 552 molecules in the asymmetric unit dock to AioAB in a cleft at the interface between the AioA and AioB subunits, with an edge-to-edge distance of 7.5 Å between the heme of cytc 552 and the [2Fe-2S] Rieske cluster in the AioB subunit. The interface between the AioAB and cytc 552 proteins features electrostatic and nonpolar interactions and is stabilized by two salt bridges. A modest number of hydrogen bonds, salt bridges and relatively small, buried surface areas between protein partners are typical features of transient electron transfer complexes. Interestingly, the fourth cytc 552 molecule is positioned differently between two AioAB heterodimers, with distances between its heme and the AioAB redox active cofactors that are outside the acceptable range for fast electron transfer. This unique cytc 552 molecule appears to be positioned to facilitate crystal packing rather than reflecting a functional complex.
- School of Chemistry and The Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Parkville, Australia.
Organizational Affiliation: