"Stapling" scFv for multispecific biotherapeutics of superior properties.
Boucher, L.E., Prinslow, E.G., Feldkamp, M., Yi, F., Nanjunda, R., Wu, S.J., Liu, T., Lacy, E.R., Jacobs, S., Kozlyuk, N., Del Rosario, B., Wu, B., Aquino, P., Davidson, R.C., Heyne, S., Mazzanti, N., Testa, J., Diem, M.D., Gorre, E., Mahan, A., Nanda, H., Gunawardena, H.P., Gervais, A., Armstrong, A.A., Teplyakov, A., Huang, C., Zwolak, A., Chowdhury, P., Cheung, W.C., Luo, J.(2023) MAbs 15: 2195517-2195517
- PubMed: 37074212
- DOI: https://doi.org/10.1080/19420862.2023.2195517
- Primary Citation of Related Structures:
8DY0, 8DY1, 8DY2, 8DY3, 8DY4, 8DY5 - PubMed Abstract:
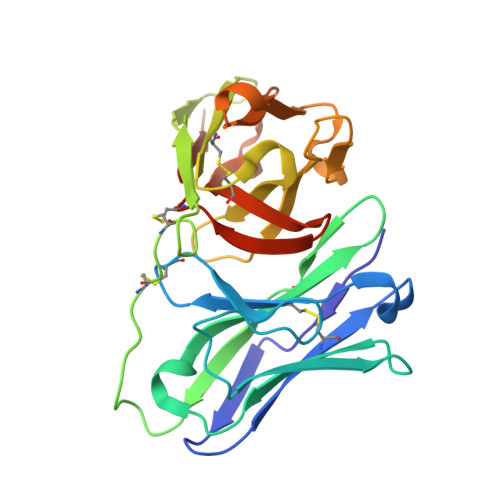
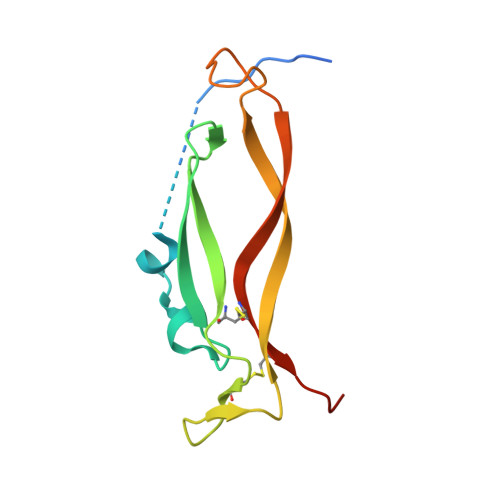
Single-chain fragment variable (scFv) domains play an important role in antibody-based therapeutic modalities, such as bispecifics, multispecifics and chimeric antigen receptor T cells or natural killer cells. However, scFv domains exhibit lower stability and increased risk of aggregation due to transient dissociation ("breathing") and inter-molecular reassociation of the two domains (VL and VH). We designed a novel strategy, referred to as stapling, that introduces two disulfide bonds between the scFv linker and the two variable domains to minimize scFv breathing. We named the resulting molecules stapled scFv (spFv). Stapling increased thermal stability (Tm) by an average of 10°C. In multiple scFv/spFv multispecifics, the spFv molecules display significantly improved stability, minimal aggregation and superior product quality. These spFv multispecifics retain binding affinity and functionality. Our stapling design was compatible with all antibody variable regions we evaluated and may be widely applicable to stabilize scFv molecules for designing biotherapeutics with superior biophysical properties.
- AbbVie, 1 North Waukegan Rd, North Chicago, IL, USA.
Organizational Affiliation: