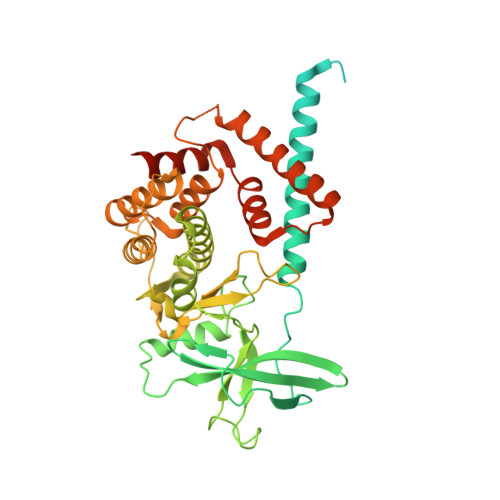
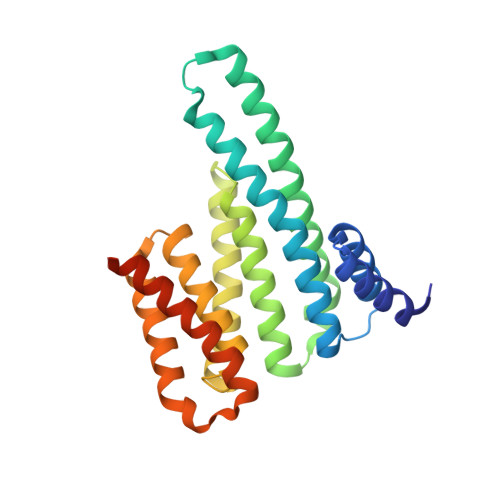
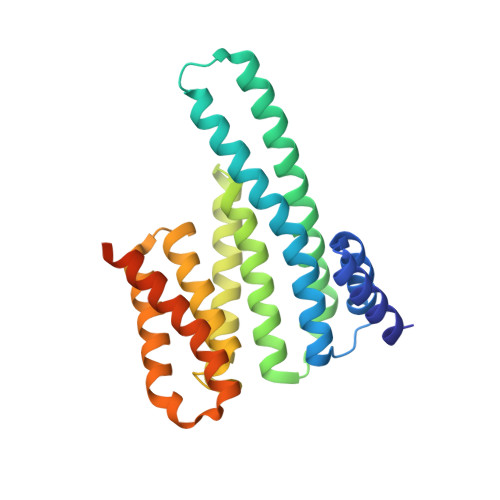
Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Torosyan, H., Paul, M.D., Forget, A., Lo, M., Diwanji, D., Pawlowski, K., Krogan, N.J., Jura, N., Verba, K.A.(2023) Nat Commun 14: 3543-3543
- PubMed: 37336883
- DOI: https://doi.org/10.1038/s41467-023-38864-0
- Primary Citation of Related Structures:
8DP5, 8DS6 - PubMed Abstract:
PEAK pseudokinases are molecular scaffolds which dimerize to regulate cell migration, morphology, and proliferation, as well as cancer progression. The mechanistic role dimerization plays in PEAK scaffolding remains unclear, as there are no structures of PEAKs in complex with their interactors. Here, we report the cryo-EM structure of dimeric PEAK3 in complex with an endogenous 14-3-3 heterodimer. Our structure reveals an asymmetric binding mode between PEAK3 and 14-3-3 stabilized by one pseudokinase domain and the SHED domain of the PEAK3 dimer. The binding interface contains a canonical phosphosite-dependent primary interaction and a unique secondary interaction not observed in previous structures of 14-3-3/client complexes. Additionally, we show that PKD regulates PEAK3/14-3-3 binding, which when prevented leads to PEAK3 nuclear enrichment and distinct protein-protein interactions. Altogether, our data demonstrate that PEAK3 dimerization forms an unusual secondary interface for 14-3-3 binding, facilitating 14-3-3 regulation of PEAK3 localization and interactome diversity.
- Cardiovascular Research Institute, University of California San Francisco, San Francisco, CA, 94158, USA.
Organizational Affiliation: