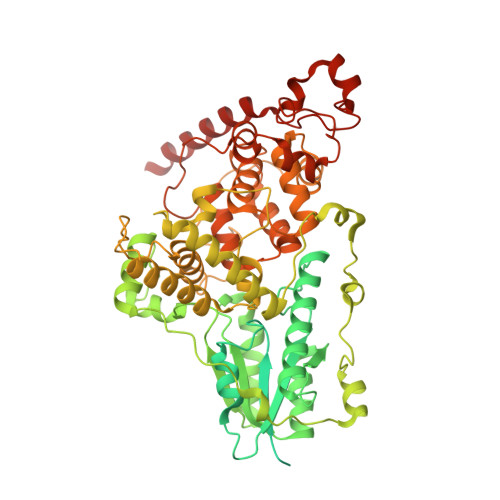
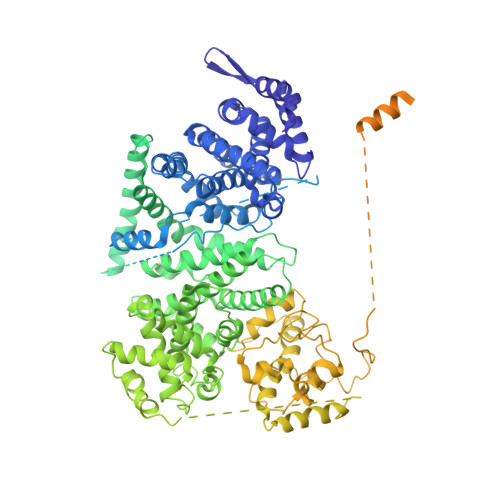
Cryptochrome-Timeless structure reveals circadian clock timing mechanisms.
Lin, C., Feng, S., DeOliveira, C.C., Crane, B.R.(2023) Nature 617: 194-199
- PubMed: 37100907
- DOI: https://doi.org/10.1038/s41586-023-06009-4
- Primary Citation of Related Structures:
8DD7 - PubMed Abstract:
Circadian rhythms influence many behaviours and diseases 1,2 . They arise from oscillations in gene expression caused by repressor proteins that directly inhibit transcription of their own genes. The fly circadian clock offers a valuable model for studying these processes, wherein Timeless (Tim) plays a critical role in mediating nuclear entry of the transcriptional repressor Period (Per) and the photoreceptor Cryptochrome (Cry) entrains the clock by triggering Tim degradation in light 2,3 . Here, through cryogenic electron microscopy of the Cry-Tim complex, we show how a light-sensing cryptochrome recognizes its target. Cry engages a continuous core of amino-terminal Tim armadillo repeats, resembling how photolyases recognize damaged DNA, and binds a C-terminal Tim helix, reminiscent of the interactions between light-insensitive cryptochromes and their partners in mammals. The structure highlights how the Cry flavin cofactor undergoes conformational changes that couple to large-scale rearrangements at the molecular interface, and how a phosphorylated segment in Tim may impact clock period by regulating the binding of Importin-α and the nuclear import of Tim-Per 4,5 . Moreover, the structure reveals that the N terminus of Tim inserts into the restructured Cry pocket to replace the autoinhibitory C-terminal tail released by light, thereby providing a possible explanation for how the long-short Tim polymorphism adapts flies to different climates 6,7 .
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY, USA.
Organizational Affiliation: