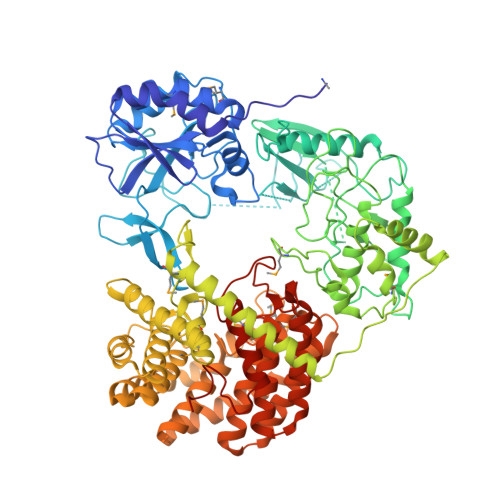
Discovery of the Lanthipeptide Curvocidin and Structural Insights into its Trifunctional Synthetase CuvL.
Sigurdsson, A., Martins, B.M., Duttmann, S.A., Jasyk, M., Dimos-Rohl, B., Schopf, F., Gemander, M., Knittel, C.H., Schnegotzki, R., Schmid, B., Kosol, S., Pommerening, L., Gonzales-Viegaz, M., Seidel, M., Hugelland, M., Leimkuhler, S., Dobbek, H., Mainz, A., Sussmuth, R.D.(2023) Angew Chem Int Ed Engl 62: e202302490-e202302490
- PubMed: 37014271
- DOI: https://doi.org/10.1002/anie.202302490
- Primary Citation of Related Structures:
8CAR, 8CAV - PubMed Abstract:
Lanthipeptides are ribosomally-synthesized natural products from bacteria featuring stable thioether-crosslinks and various bioactivities. Herein, we report on a new clade of tricyclic class-IV lanthipeptides with curvocidin from Thermomonospora curvata as its first representative. We obtained crystal structures of the corresponding lanthipeptide synthetase CuvL that showed a circular arrangement of its kinase, lyase and cyclase domains, forming a central reaction chamber for the iterative substrate processing involving nine catalytic steps. The combination of experimental data and artificial intelligence-based structural models identified the N-terminal subdomain of the kinase domain as the primary site of substrate recruitment. The ribosomal precursor peptide of curvocidin employs an amphipathic α-helix in its leader region as an anchor to CuvL, while its substrate core shuttles within the central reaction chamber. Our study thus reveals general principles of domain organization and substrate recruitment of class-IV and class-III lanthipeptide synthetases.
- Fakultät II-Institut für Chemie, Technische Universität Berlin, Straße des 17. Juni 124, 10623, Berlin, Germany.
Organizational Affiliation: