Preclinical anti-tumour activity of HexaBody-CD38, a next-generation CD38 antibody with superior complement-dependent cytotoxic activity.
Hiemstra, I.H., Santegoets, K.C.M., Janmaat, M.L., De Goeij, B.E.C.G., Ten Hagen, W., van Dooremalen, S., Boross, P., van den Brakel, J., Bosgra, S., Andringa, G., van Kessel-Welmers, B., Verzijl, D., Hibbert, R.G., Frerichs, K.A., Mutis, T., van de Donk, N.W.C.J., Ahmadi, T., Satijn, D., Sasser, A.K., Breij, E.C.W.(2023) EBioMedicine 93: 104663-104663
- PubMed: 37379657
- DOI: https://doi.org/10.1016/j.ebiom.2023.104663
- Primary Citation of Related Structures:
8BYU - PubMed Abstract:
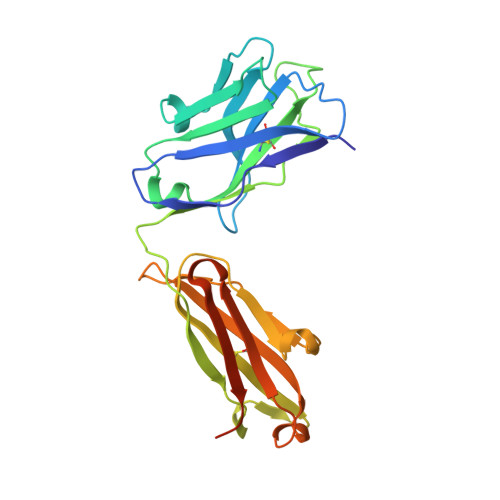
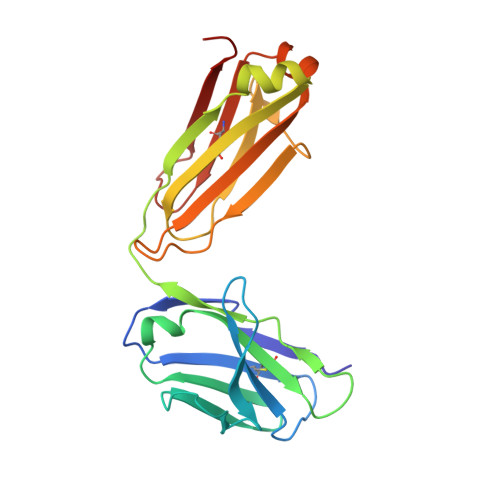
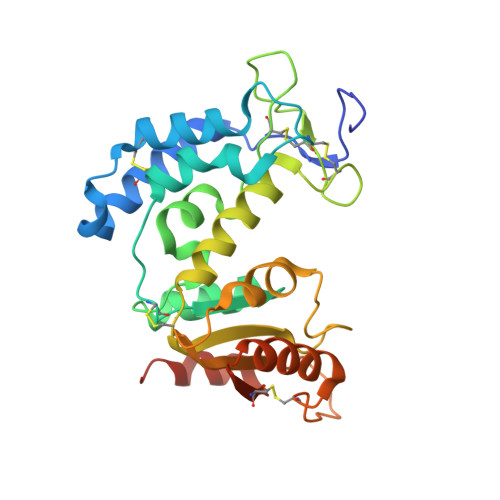
HexaBody®-CD38 (GEN3014) is a hexamerization-enhanced human IgG1 that binds CD38 with high affinity. The E430G mutation in its Fc domain facilitates the natural process of antibody hexamer formation upon binding to the cell surface, resulting in increased binding of C1q and potentiated complement-dependent cytotoxicity (CDC). Co-crystallization studies were performed to identify the binding interface of HexaBody-CD38 and CD38. HexaBody-CD38-induced CDC, antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), trogocytosis, and apoptosis were assessed using flow cytometry assays using tumour cell lines, and MM patient samples (CDC). CD38 enzymatic activity was measured using fluorescence spectroscopy. Anti-tumour activity of HexaBody-CD38 was assessed in patient-derived xenograft mouse models in vivo. HexaBody-CD38 binds a unique epitope on CD38 and induced potent CDC in multiple myeloma (MM), acute myeloid leukaemia (AML), and B-cell non-Hodgkin lymphoma (B-NHL) cells. Anti-tumour activity was confirmed in patient-derived xenograft models in vivo. Sensitivity to HexaBody-CD38 correlated with CD38 expression level and was inversely correlated with expression of complement regulatory proteins. Compared to daratumumab, HexaBody-CD38 showed enhanced CDC in cell lines with lower levels of CD38 expression, without increasing lysis of healthy leukocytes. More effective CDC was also confirmed in primary MM cells. Furthermore, HexaBody-CD38 efficiently induced ADCC, ADCP, trogocytosis, and apoptosis after Fc-crosslinking. Moreover, HexaBody-CD38 strongly inhibited CD38 cyclase activity, which is hypothesized to relieve immune suppression in the tumour microenvironment. Based on these preclinical studies, a clinical trial was initiated to assess the clinical safety of HexaBody-CD38 in patients with MM. Genmab.
- Genmab B.V., Utrecht, the Netherlands.
Organizational Affiliation: