Crystal structure of the phage Mu protein Mom catalytic mutant S124A
Silva, R.M.B., Slyvka, A., Lee, Y.J., Guan, C., Lund, S.R., Raleigh, E.A., Skowronek, K., Bochtler, M., Weigele, P.R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methylcarbamoylase mom | 232 | Escherichia phage Mu | Mutation(s): 1 Gene Names: mom, Mup55 | 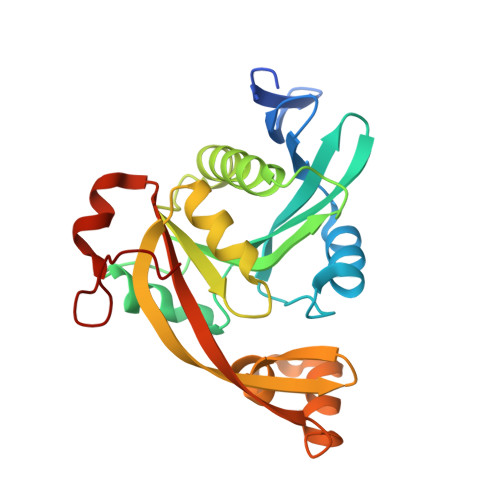 | |
UniProt | |||||
Find proteins for P06018 (Escherichia phage Mu) Explore P06018 Go to UniProtKB: P06018 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06018 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.18 | α = 90 |
| b = 66.48 | β = 90 |
| c = 138.39 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| PHENIX | phasing |
| Coot | model building |
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Foundation for Polish Science | Poland | POIR.04.04.00-00-5D81/17-00 |
| Polish National Science Centre | Poland | 2018/30/Q/NZ2/00669 |
| Other government | Polish National Agency for Academic Exchange PPI/APM/2018/1/00034 | |
| Other private | New England Biolabs |