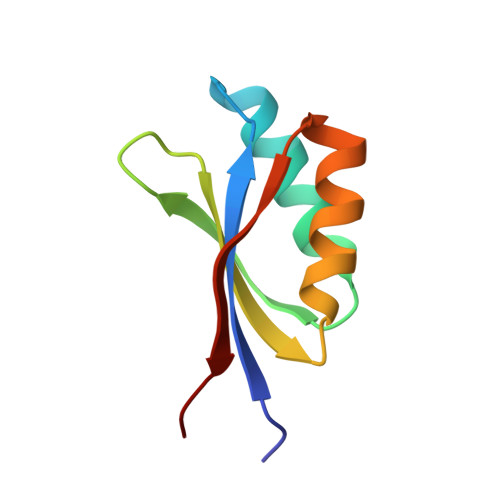
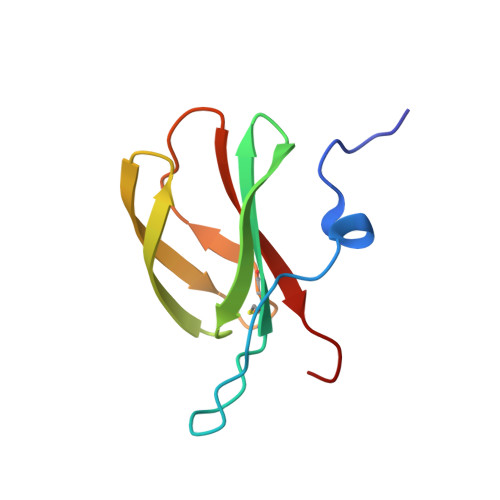
Allelic compatibility in plant immune receptors facilitates engineering of new effector recognition specificities.
Bentham, A.R., De la Concepcion, J.C., Benjumea, J.V., Kourelis, J., Jones, S., Mendel, M., Stubbs, J., Stevenson, C.E.M., Maidment, J.H.R., Youles, M., Zdrzalek, R., Kamoun, S., Banfield, M.J.(2023) Plant Cell 35: 3809-3827
- PubMed: 37486356
- DOI: https://doi.org/10.1093/plcell/koad204
- Primary Citation of Related Structures:
8B2R - PubMed Abstract:
Engineering the plant immune system offers genetic solutions to mitigate crop diseases caused by diverse agriculturally significant pathogens and pests. Modification of intracellular plant immune receptors of the nucleotide-binding leucine-rich repeat (NLR) receptor superfamily for expanded recognition of pathogen virulence proteins (effectors) is a promising approach for engineering disease resistance. However, engineering can cause NLR autoactivation, resulting in constitutive defense responses that are deleterious to the plant. This may be due to plant NLRs associating in highly complex signaling networks that coevolve together, and changes through breeding or genetic modification can generate incompatible combinations, resulting in autoimmune phenotypes. The sensor and helper NLRs of the rice (Oryza sativa) NLR pair Pik have coevolved, and mismatching between noncoevolved alleles triggers constitutive activation and cell death. This limits the extent to which protein modifications can be used to engineer pathogen recognition and enhance disease resistance mediated by these NLRs. Here, we dissected incompatibility determinants in the Pik pair in Nicotiana benthamiana and found that heavy metal-associated (HMA) domains integrated in Pik-1 not only evolved to bind pathogen effectors but also likely coevolved with other NLR domains to maintain immune homeostasis. This explains why changes in integrated domains can lead to autoactivation. We then used this knowledge to facilitate engineering of new effector recognition specificities, overcoming initial autoimmune penalties. We show that by mismatching alleles of the rice sensor and helper NLRs Pik-1 and Pik-2, we can enable the integration of synthetic domains with novel and enhanced recognition specificities. Taken together, our results reveal a strategy for engineering NLRs, which has the potential to allow an expanded set of integrations and therefore new disease resistance specificities in plants.
- Department of Biochemistry and Metabolism, John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK.
Organizational Affiliation: