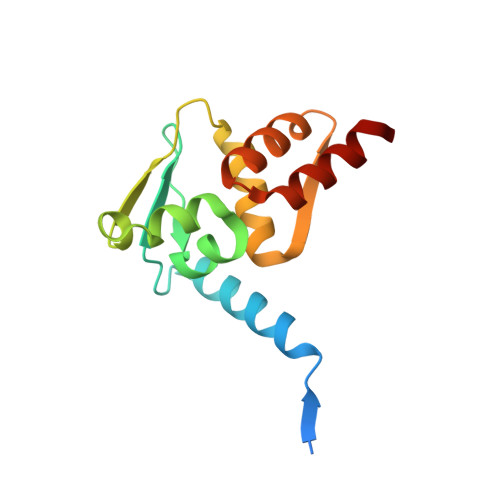
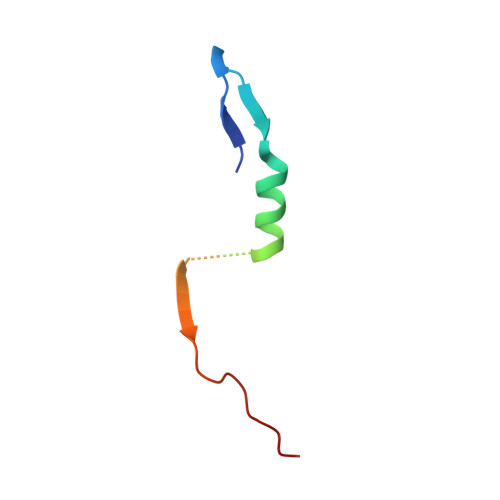
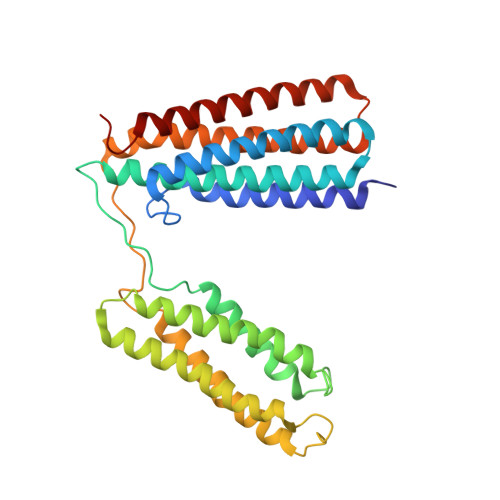
The structural basis of the talin-KANK1 interaction that coordinates the actin and microtubule cytoskeletons at focal adhesions.
Li, X., Goult, B.T., Ballestrem, C., Zacharchenko, T.(2023) Open Biol 13: 230058-230058
- PubMed: 37339751
- DOI: https://doi.org/10.1098/rsob.230058
- Primary Citation of Related Structures:
8AS9 - PubMed Abstract:
Adhesion between cells and the extracellular matrix is mediated by heterodimeric ( αβ ) integrin receptors that are intracellularly linked to the contractile actomyosin machinery. One of the proteins that control this link is talin, which organizes cytosolic signalling proteins into discrete complexes on β-integrin tails referred to as focal adhesions (FAs). The adapter protein KANK1 binds to talin in the region of FAs known as the adhesion belt. Here, we adapted a non-covalent crystallographic chaperone to resolve the talin-KANK1 complex. This structure revealed that the talin binding KN region of KANK1 contains a novel motif where a β-hairpin stabilizes the α-helical region, explaining both its specific interaction with talin R7 and high affinity. Single point mutants in KANK1 identified from the structure abolished the interaction and enabled us to examine KANK1 enrichment in the adhesion belt. Strikingly, in cells expressing a constitutively active form of vinculin that keeps the FA structure intact even in the presence of myosin inhibitors, KANK1 localizes throughout the entire FA structure even when actomyosin tension is released. We propose a model whereby actomyosin forces on talin eliminate KANK1 from talin binding in the centre of FAs while retaining it at the adhesion periphery.
- Wellcome Centre for Cell-Matrix Research, Faculty of Biology, Medicine and Health, University of Manchester, Dover Street, Manchester M13 9PT, UK.
Organizational Affiliation: