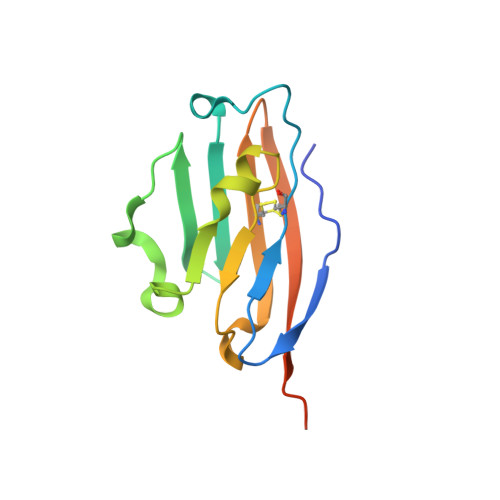
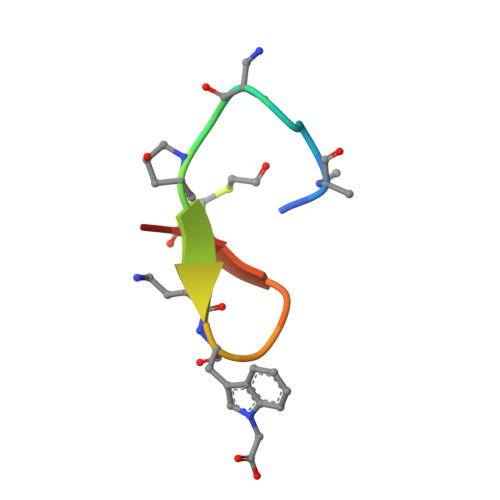
Structural and biological characterization of pAC65, a macrocyclic peptide that blocks PD-L1 with equivalent potency to the FDA-approved antibodies.
Rodriguez, I., Kocik-Krol, J., Skalniak, L., Musielak, B., Wisniewska, A., Ciesiolkiewicz, A., Berlicki, L., Plewka, J., Grudnik, P., Stec, M., Siedlar, M., Holak, T.A., Magiera-Mularz, K.(2023) Mol Cancer 22: 150-150
- PubMed: 37679783
- DOI: https://doi.org/10.1186/s12943-023-01853-4
- Primary Citation of Related Structures:
8ALX - PubMed Abstract:
Recent advances in immuno-oncology have opened up new and impressive treatment options for cancer. Notwithstanding, overcoming the limitations of the current FDA-approved therapies with monoclonal antibodies (mAbs) that block the PD-1/PD-L1 pathway continues to lead to the testing of multiple approaches and optimizations. Recently, a series of macrocyclic peptides have been developed that exhibit binding strengths to PD-L1 ranging from sub-micromolar to micromolar. In this study, we present the most potent non-antibody-based PD-1/PD-L1 interaction inhibitor reported to date. The structural and biological characterization of this macrocyclic PD-L1 targeting peptide provides the rationale for inhibition of both PD-1/PD-L1 and CD80/PD-L1 complexes. The IC 50 and EC 50 values obtained in PD-L1 binding assays indicate that the pAC65 peptide has potency equivalent to the current FDA-approved mAbs and may have similar activity to the BMS986189 peptide, which entered the clinical trial and has favorable safety and pharmacokinetic data. The data presented here delineate the generation of similar peptides with improved biological activities and applications not only in the field of cancer immunotherapy but also in other disorders related to the immune system.
- Department of Organic Chemistry, Faculty of Chemistry, Jagiellonian University, Gronostajowa 2, Krakow, 30-387, Poland.
Organizational Affiliation: