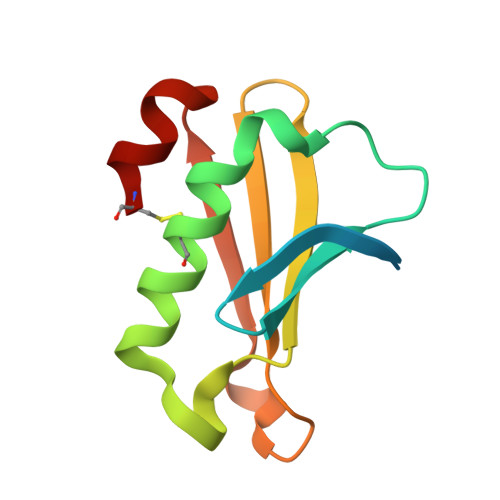
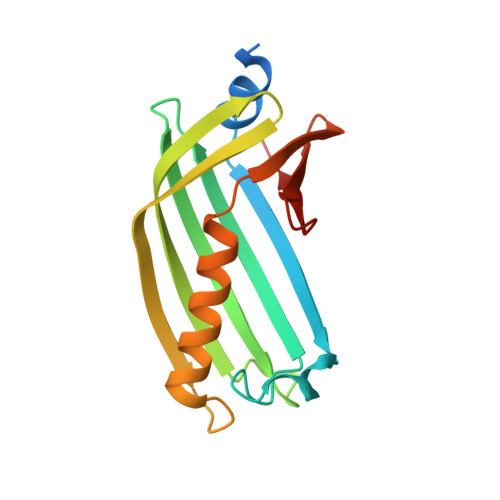
Vibrio cholerae's ToxRS bile sensing system.
Gubensak, N., Sagmeister, T., Buhlheller, C., Geronimo, B.D., Wagner, G.E., Petrowitsch, L., Grawert, M.A., Rotzinger, M., Berger, T.M.I., Schafer, J., Uson, I., Reidl, J., Sanchez-Murcia, P.A., Zangger, K., Pavkov-Keller, T.(2023) Elife 12
- PubMed: 37768326
- DOI: https://doi.org/10.7554/eLife.88721
- Primary Citation of Related Structures:
8ALO - PubMed Abstract:
The seventh pandemic of the diarrheal cholera disease, which began in 1960, is caused by the Gram-negative bacterium Vibrio cholerae . Its environmental persistence provoking recurring sudden outbreaks is enabled by V. cholerae's rapid adaption to changing environments involving sensory proteins like ToxR and ToxS. Located at the inner membrane, ToxR and ToxS react to environmental stimuli like bile acid, thereby inducing survival strategies for example bile resistance and virulence regulation. The presented crystal structure of the sensory domains of ToxR and ToxS in combination with multiple bile acid interaction studies, reveals that a bile binding pocket of ToxS is only properly folded upon binding to ToxR. Our data proposes an interdependent functionality between ToxR transcriptional activity and ToxS sensory function. These findings support the previously suggested link between ToxRS and VtrAC-like co-component systems. Besides VtrAC, ToxRS is now the only experimentally determined structure within this recently defined superfamily, further emphasizing its significance. In-depth analysis of the ToxRS complex reveals its remarkable conservation across various Vibrio species, underlining the significance of conserved residues in the ToxS barrel and the more diverse ToxR sensory domain. Unravelling the intricate mechanisms governing ToxRS's environmental sensing capabilities, provides a promising tool for disruption of this vital interaction, ultimately inhibiting Vibrio's survival and virulence. Our findings hold far-reaching implications for all Vibrio strains that rely on the ToxRS system as a shared sensory cornerstone for adapting to their surroundings.
- Institute of Molecular Biosciences, University of Graz, Graz, Austria.
Organizational Affiliation: