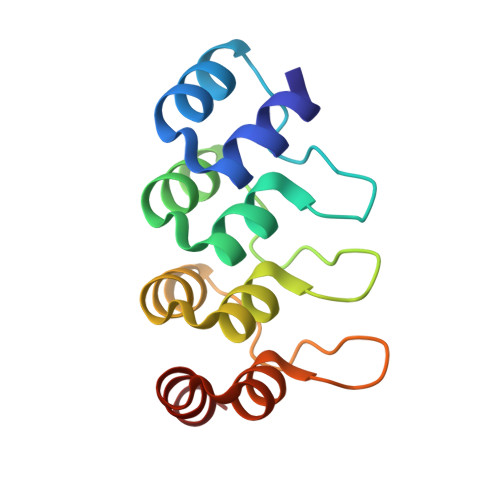
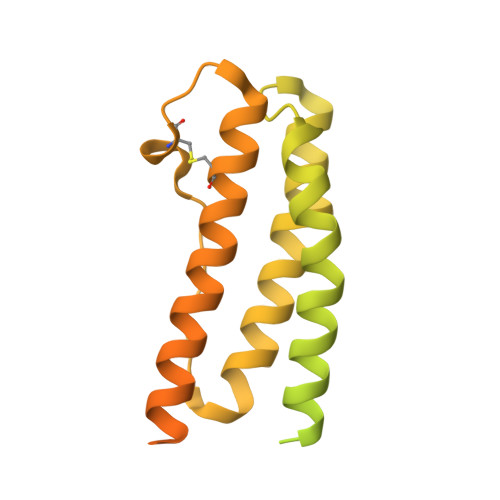
Designed Ankyrin Repeat Proteins provide insights into the structure and function of CagI and are potent inhibitors of CagA translocation by the Helicobacter pylori type IV secretion system.
Blanc, M., Lettl, C., Guerin, J., Vieille, A., Furler, S., Briand-Schumacher, S., Dreier, B., Berge, C., Pluckthun, A., Vadon-Le Goff, S., Fronzes, R., Rousselle, P., Fischer, W., Terradot, L.(2023) PLoS Pathog 19: e1011368-e1011368
- PubMed: 37155700
- DOI: https://doi.org/10.1371/journal.ppat.1011368
- Primary Citation of Related Structures:
8AIW, 8AK1 - PubMed Abstract:
The bacterial human pathogen Helicobacter pylori produces a type IV secretion system (cagT4SS) to inject the oncoprotein CagA into gastric cells. The cagT4SS external pilus mediates attachment of the apparatus to the target cell and the delivery of CagA. While the composition of the pilus is unclear, CagI is present at the surface of the bacterium and required for pilus formation. Here, we have investigated the properties of CagI by an integrative structural biology approach. Using Alpha Fold 2 and Small Angle X-ray scattering, it was found that CagI forms elongated dimers mediated by rod-shape N-terminal domains (CagIN) prolonged by globular C-terminal domains (CagIC). Three Designed Ankyrin Repeat Proteins (DARPins) K2, K5 and K8 selected against CagI interacted with CagIC with subnanomolar affinities. The crystal structures of the CagI:K2 and CagI:K5 complexes were solved and identified the interfaces between the molecules, thereby providing a structural explanation for the difference in affinity between the two binders. Purified CagI and CagIC were found to interact with adenocarcinoma gastric (AGS) cells, induced cell spreading and the interaction was inhibited by K2. The same DARPin inhibited CagA translocation by up to 65% in AGS cells while inhibition levels were 40% and 30% with K8 and K5, respectively. Our study suggests that CagIC plays a key role in cagT4SS-mediated CagA translocation and that DARPins targeting CagI represent potent inhibitors of the cagT4SS, a crucial risk factor for gastric cancer development.
- UMR 5086 Molecular Microbiology and Structural Biochemistry CNRS-Université de Lyon, Institut de Biologie et Chimie des Protéines, Lyon, France.
Organizational Affiliation: