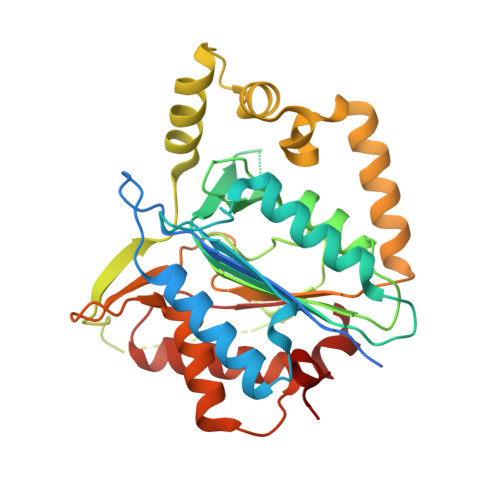
Structure-function study of a Ca 2+ -independent metacaspase involved in lateral root emergence.
Stael, S., Sabljic, I., Audenaert, D., Andersson, T., Tsiatsiani, L., Kumpf, R.P., Vidal-Albalat, A., Lindgren, C., Vercammen, D., Jacques, S., Nguyen, L., Njo, M., Fernandez-Fernandez, A.D., Beunens, T., Timmerman, E., Gevaert, K., Van Montagu, M., Stahlberg, J., Bozhkov, P.V., Linusson, A., Beeckman, T., Van Breusegem, F.(2023) Proc Natl Acad Sci U S A 120: e2303480120-e2303480120
- PubMed: 37216519
- DOI: https://doi.org/10.1073/pnas.2303480120
- Primary Citation of Related Structures:
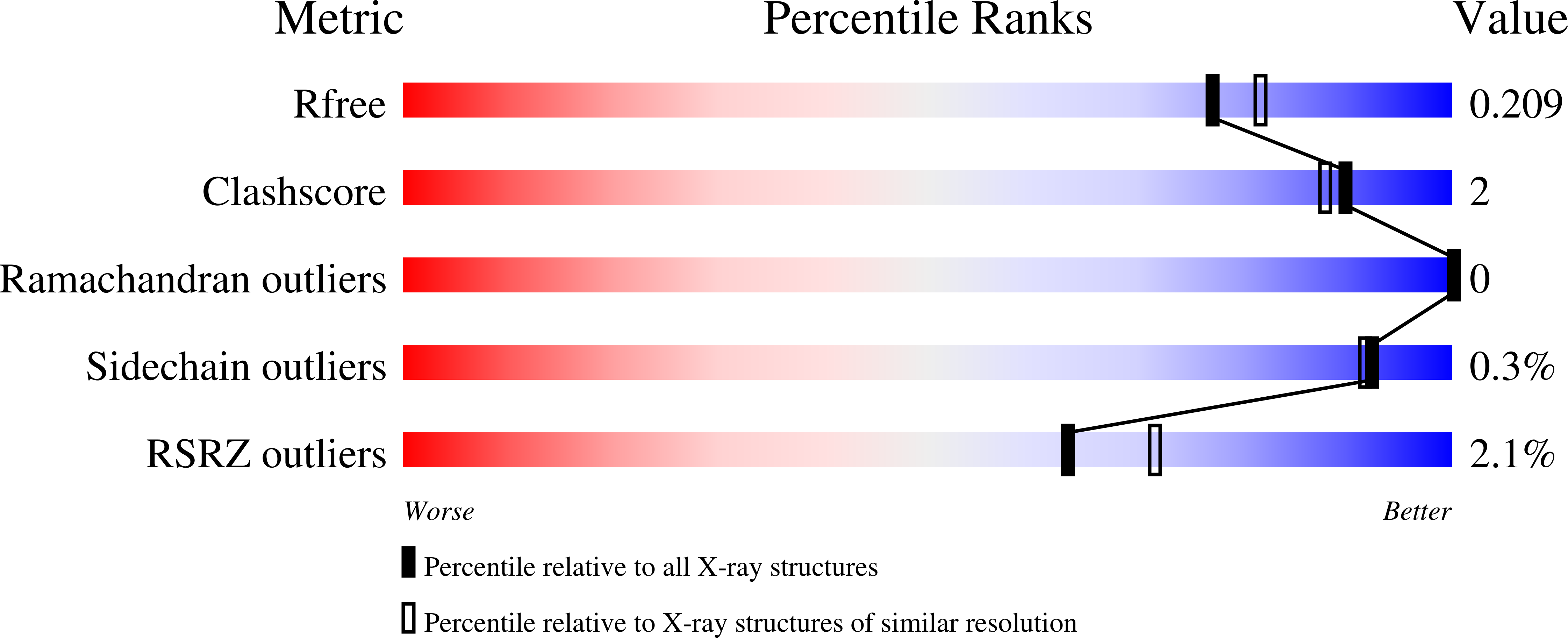
8A53 - PubMed Abstract:
Metacaspases are part of an evolutionarily broad family of multifunctional cysteine proteases, involved in disease and normal development. As the structure-function relationship of metacaspases remains poorly understood, we solved the X-ray crystal structure of an Arabidopsis thaliana type II metacaspase (AtMCA-IIf) belonging to a particular subgroup not requiring calcium ions for activation. To study metacaspase activity in plants, we developed an in vitro chemical screen to identify small molecule metacaspase inhibitors and found several hits with a minimal thioxodihydropyrimidine-dione structure, of which some are specific AtMCA-IIf inhibitors. We provide mechanistic insight into the basis of inhibition by the TDP-containing compounds through molecular docking onto the AtMCA-IIf crystal structure. Finally, a TDP-containing compound (TDP6) effectively hampered lateral root emergence in vivo, probably through inhibition of metacaspases specifically expressed in the endodermal cells overlying developing lateral root primordia. In the future, the small compound inhibitors and crystal structure of AtMCA-IIf can be used to study metacaspases in other species, such as important human pathogens, including those causing neglected diseases.
- Department of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Ghent, Belgium.
Organizational Affiliation: