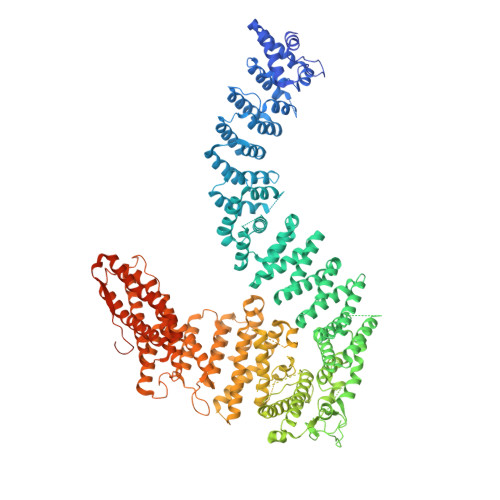
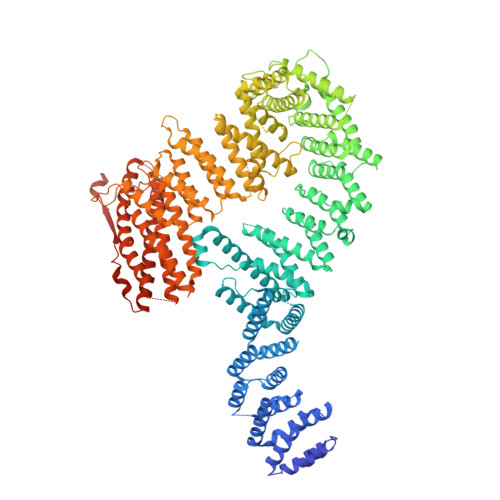
The DNA-damage kinase ATR activates the FANCD2-FANCI clamp by priming it for ubiquitination.
Sijacki, T., Alcon, P., Chen, Z.A., McLaughlin, S.H., Shakeel, S., Rappsilber, J., Passmore, L.A.(2022) Nat Struct Mol Biol 29: 881-890
- PubMed: 36050501
- DOI: https://doi.org/10.1038/s41594-022-00820-9
- Primary Citation of Related Structures:
8A2Q - PubMed Abstract:
DNA interstrand cross-links are tumor-inducing lesions that block DNA replication and transcription. When cross-links are detected at stalled replication forks, ATR kinase phosphorylates FANCI, which stimulates monoubiquitination of the FANCD2-FANCI clamp by the Fanconi anemia core complex. Monoubiquitinated FANCD2-FANCI is locked onto DNA and recruits nucleases that mediate DNA repair. However, it remains unclear how phosphorylation activates this pathway. Here, we report structures of FANCD2-FANCI complexes containing phosphomimetic FANCI. We observe that, unlike wild-type FANCD2-FANCI, the phosphomimetic complex closes around DNA, independent of the Fanconi anemia core complex. The phosphomimetic mutations do not substantially alter DNA binding but instead destabilize the open state of FANCD2-FANCI and alter its conformational dynamics. Overall, our results demonstrate that phosphorylation primes the FANCD2-FANCI clamp for ubiquitination, showing how multiple posttranslational modifications are coordinated to control DNA repair.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: