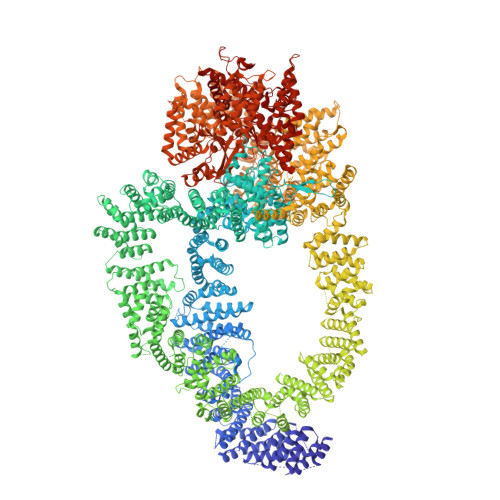
The structure of the NuA4-Tip60 complex reveals the mechanism and importance of long-range chromatin modification.
Frechard, A., Faux, C., Hexnerova, R., Crucifix, C., Papai, G., Smirnova, E., McKeon, C., Ping, F.L.Y., Helmlinger, D., Schultz, P., Ben-Shem, A.(2023) Nat Struct Mol Biol 30: 1337-1345
- PubMed: 37550452
- DOI: https://doi.org/10.1038/s41594-023-01056-x
- Primary Citation of Related Structures:
7ZVW - PubMed Abstract:
Histone acetylation regulates most DNA transactions and is dynamically controlled by highly conserved enzymes. The only essential histone acetyltransferase (HAT) in yeast, Esa1, is part of the 1-MDa NuA4 complex, which plays pivotal roles in both transcription and DNA-damage repair. NuA4 has the unique capacity to acetylate histone targets located several nucleosomes away from its recruitment site. Neither the molecular mechanism of this activity nor its physiological importance are known. Here we report the structure of the Pichia pastoris NuA4 complex, with its core resolved at 3.4-Å resolution. Three subunits, Epl1, Eaf1 and Swc4, intertwine to form a stable platform that coordinates all other modules. The HAT module is firmly anchored into the core while retaining the ability to stretch out over a long distance. We provide structural, biochemical and genetic evidence that an unfolded linker region of the Epl1 subunit is critical for this long-range activity. Specifically, shortening the Epl1 linker causes severe growth defects and reduced H4 acetylation levels over broad chromatin regions in fission yeast. Our work lays the foundations for a mechanistic understanding of NuA4's regulatory role and elucidates how its essential long-range activity is attained.
- Institut de Génétique et de Biologie Moléculaire et Cellulaire, Integrated Structural Biology Department, Illkirch, France.
Organizational Affiliation: