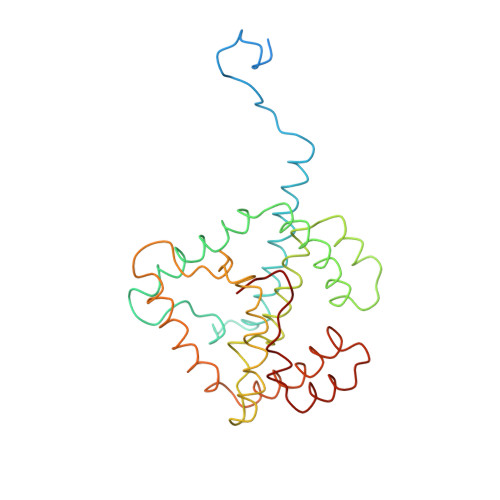
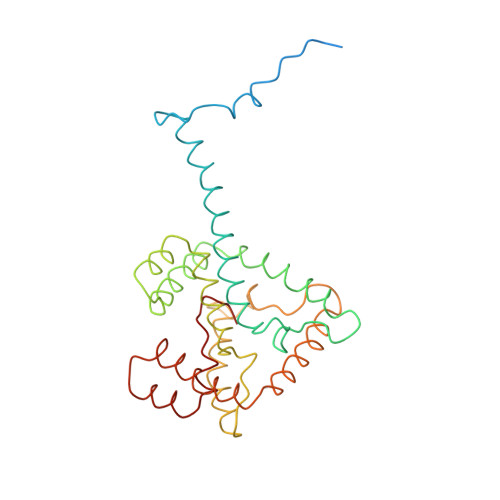
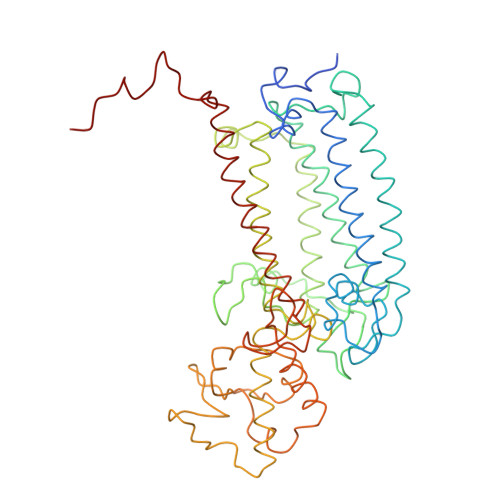
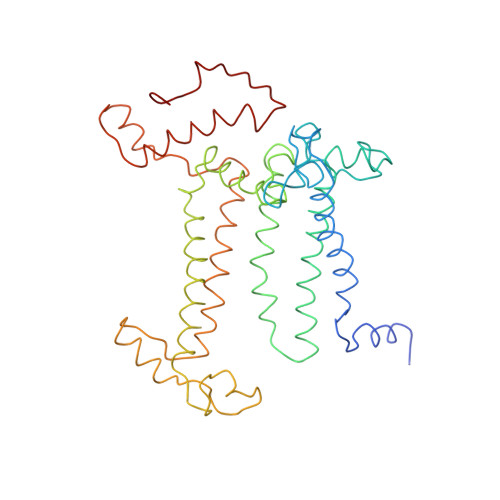
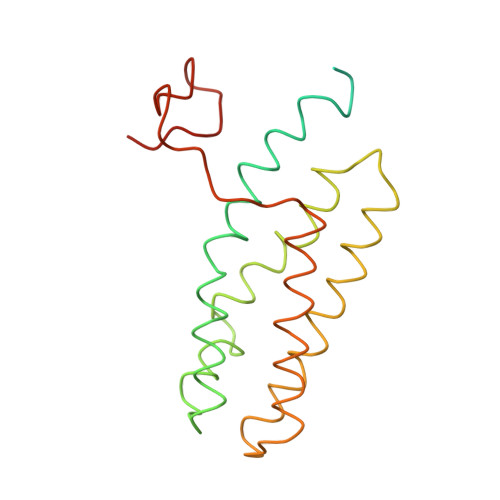
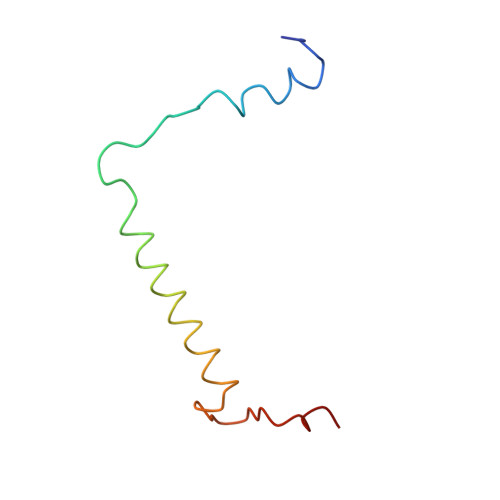
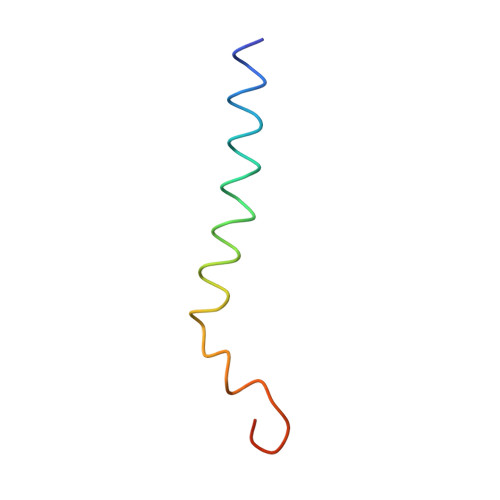
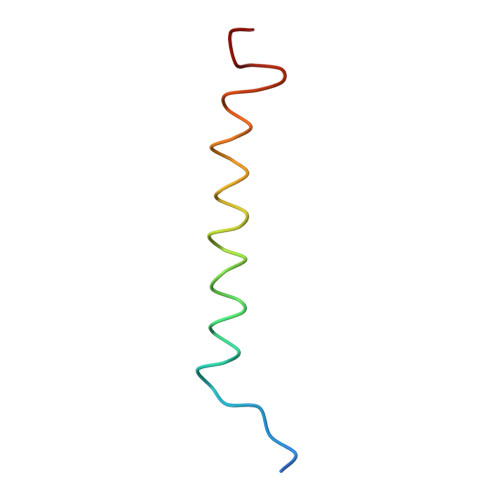
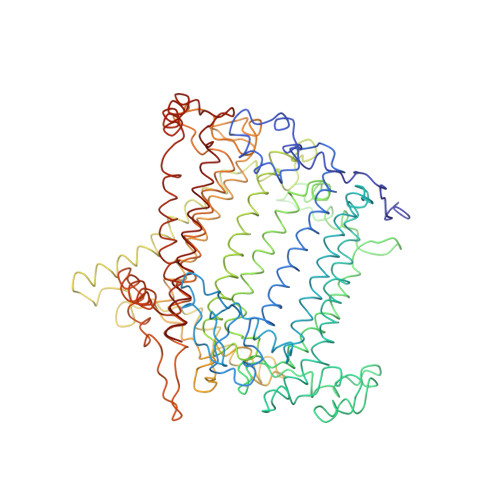
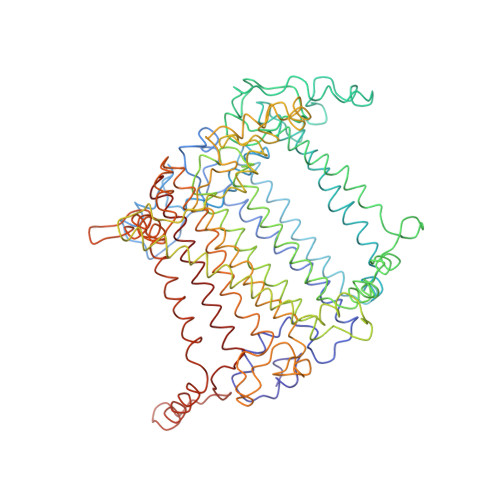
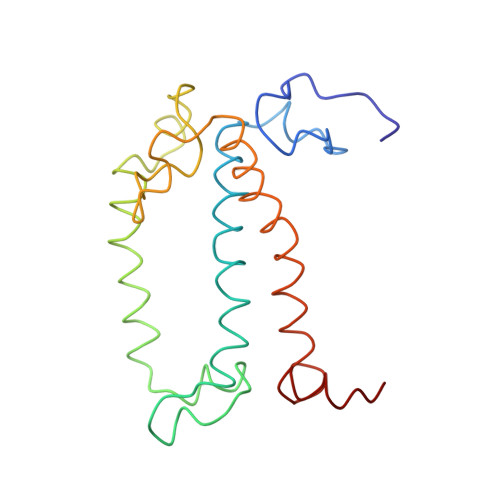
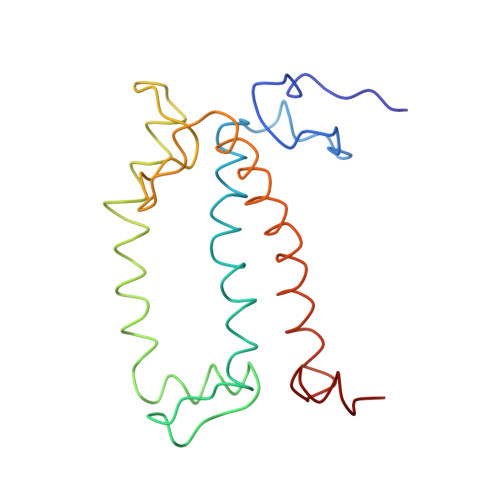
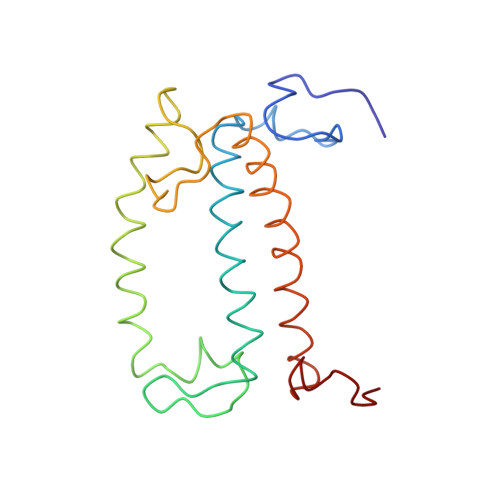
CLA Query on CLA Download Ideal Coordinates CCD File AAC [auth 32]
AAC [auth 32],
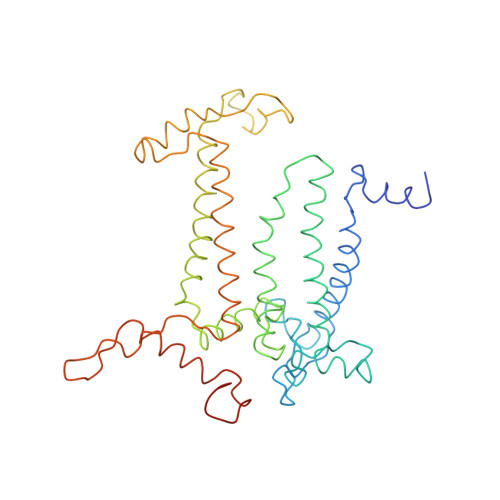
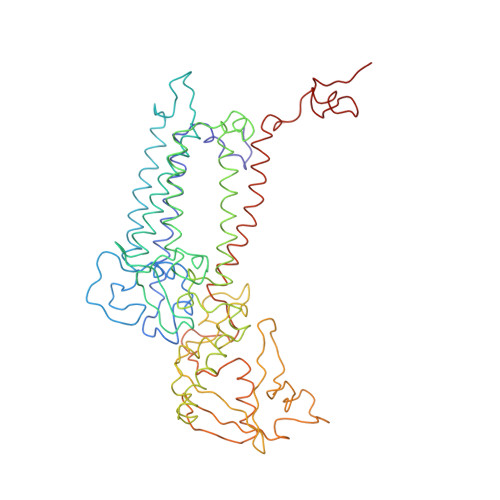
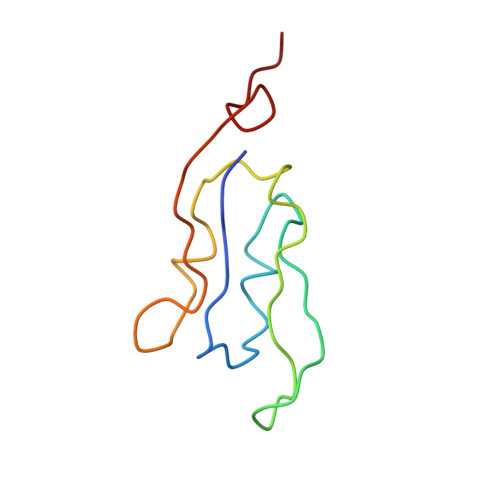
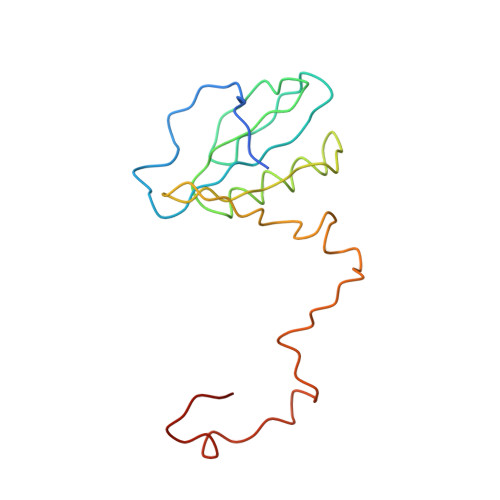
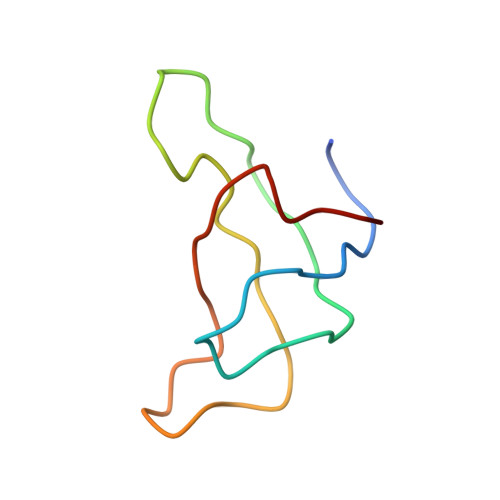
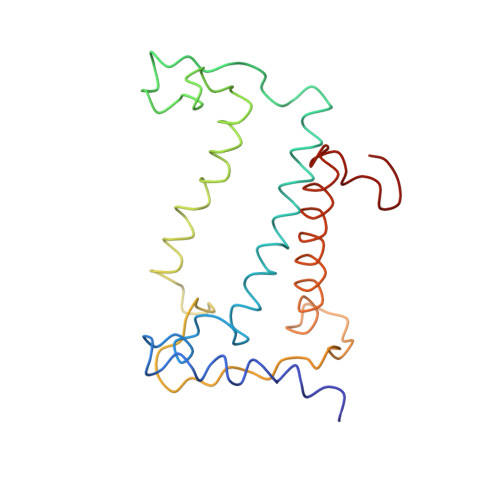
CHLOROPHYLL A 55 H72 Mg N4 O5 PEB Query on PEB Download Ideal Coordinates CCD File AAB [auth IJ]
AAB [auth IJ],
PHYCOERYTHROBILIN 33 H40 N4 O6