Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
Ikeuchi, T., Yasumoto, M., Takita, T., Tanaka, K., Kusubata, M., Hayashida, O., Hattori, S., Mizutani, K., Mikami, B., Yasukawa, K.(2022) J Biological Chem 298: 102109-102109
- PubMed: 35679897
- DOI: https://doi.org/10.1016/j.jbc.2022.102109
- Primary Citation of Related Structures:
7WSS, 7XEB - PubMed Abstract:
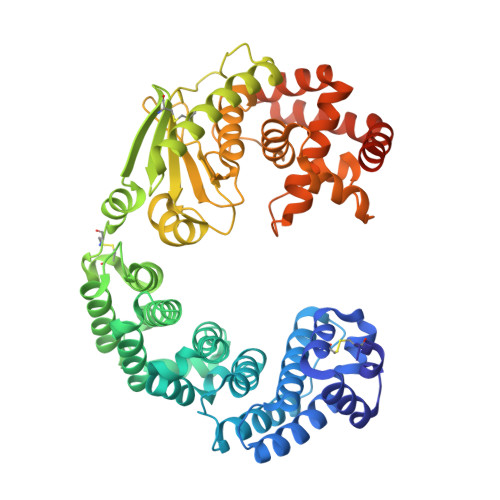
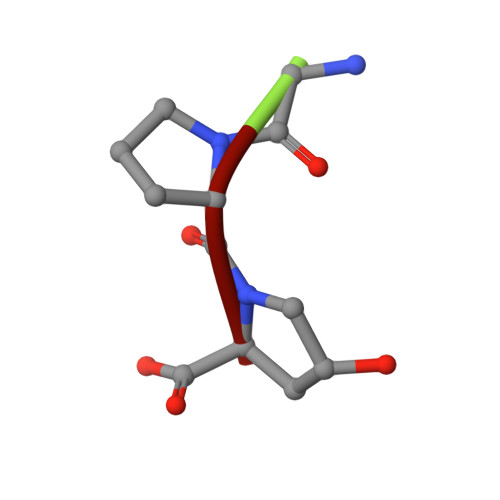
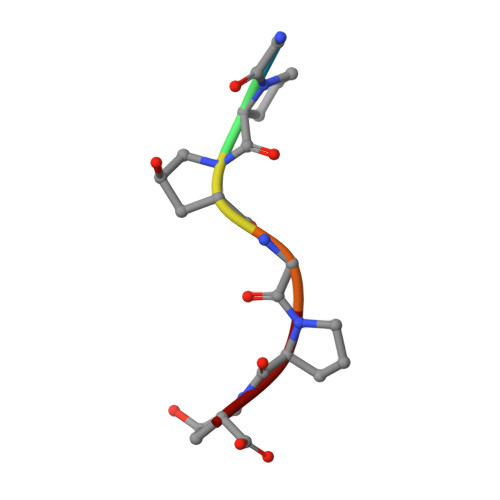
Collagenase from the gram-negative bacterium Grimontia hollisae strain 1706B (Ghcol) degrades collagen more efficiently even than clostridial collagenase, the most widely used industrial collagenase. However, the structural determinants facilitating this efficiency are unclear. Here, we report the crystal structures of ligand-free and Gly-Pro-hydroxyproline (Hyp)-complexed Ghcol at 2.2 and 2.4 Å resolution, respectively. These structures revealed that the activator and peptidase domains in Ghcol form a saddle-shaped structure with one zinc ion and four calcium ions. In addition, the activator domain comprises two homologous subdomains, whereas zinc-bound water was observed in the ligand-free Ghcol. In the ligand-complexed Ghcol, we found two Gly-Pro-Hyp molecules, each bind at the active site and at two surfaces on the duplicate subdomains of the activator domain facing the active site, and the nucleophilic water is replaced by the carboxyl oxygen of Hyp at the P1 position. Furthermore, all Gly-Pro-Hyp molecules bound to Ghcol have almost the same conformation as Pro-Pro-Gly motif in model collagen (Pro-Pro-Gly) 10 , suggesting these three sites contribute to the unwinding of the collagen triple helix. A comparison of activities revealed that Ghcol exhibits broader substrate specificity than clostridial collagenase at the P2 and P2' positions, which may be attributed to the larger space available for substrate binding at the S2 and S2' sites in Ghcol. Analysis of variants of three active-site Tyr residues revealed that mutation of Tyr564 affected catalysis, whereas mutation of Tyr476 or Tyr555 affected substrate recognition. These results provide insights into the substrate specificity and mechanism of G. hollisae collagenase.
- Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Sakyo-ku, Kyoto, Japan.
Organizational Affiliation: