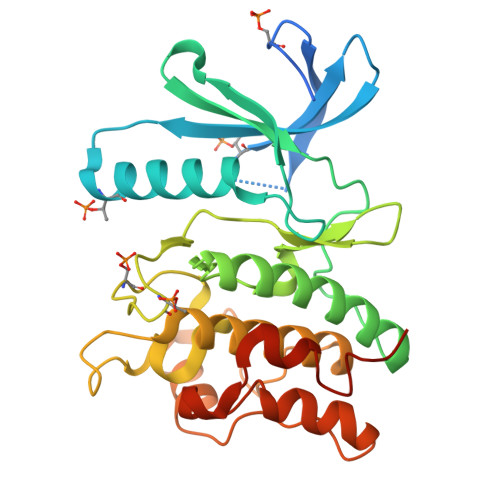
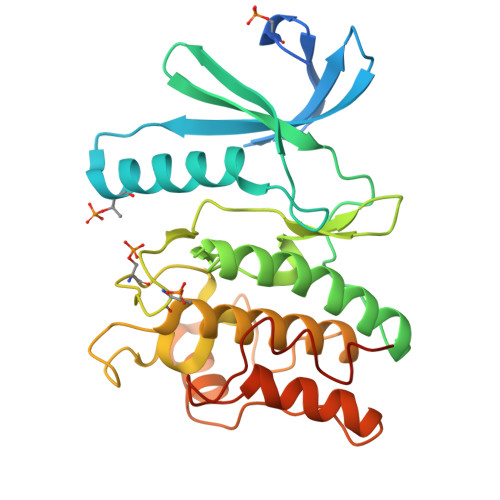
Crystal structure of the phosphorylated Arabidopsis MKK5 reveals activation mechanism of MAPK kinases.
Pei, C.J., He, Q.X., Luo, Z., Yao, H., Wang, Z.X., Wu, J.W.(2022) Acta Biochim Biophys Sin (Shanghai) 54: 1159-1170
- PubMed: 35866601
- DOI: https://doi.org/10.3724/abbs.2022089
- Primary Citation of Related Structures:
7XBR - PubMed Abstract:
The mitogen-activated protein kinase (MAPK) signaling pathways are highly conserved in eukaryotes, regulating various cellular processes. The MAPK kinases (MKKs) are dual specificity kinases, serving as convergence and divergence points of the tripartite MAPK cascades. Here, we investigate the biochemical characteristics and three-dimensional structure of MKK5 in Arabidopsis (AtMKK5). The recombinant full-length AtMKK5 is phosphorylated and can activate its physiological substrate AtMPK6. There is a conserved kinase interacting motif (KIM) at the N-terminus of AtMKK5, indispensable for specific recognition of AtMPK6. The kinase domain of AtMKK5 adopts active conformation, of which the extended activation segment is stabilized by the phosphorylated Ser221 and Thr215 residues. In line with sequence divergence from other MKKs, the αD and αK helices are missing in AtMKK5, suggesting that the AtMKK5 may adopt distinct modes of upstream kinase/substrate binding. Our data shed lights on the molecular mechanisms of MKK activation and substrate recognition, which may help design specific inhibitors targeting human and plant MKKs.
- Institute of Molecular Enzymology, School of Biology & Basic Medical Sciences, Suzhou Medical College of Soochow University, Suzhou 215123, China.
Organizational Affiliation: