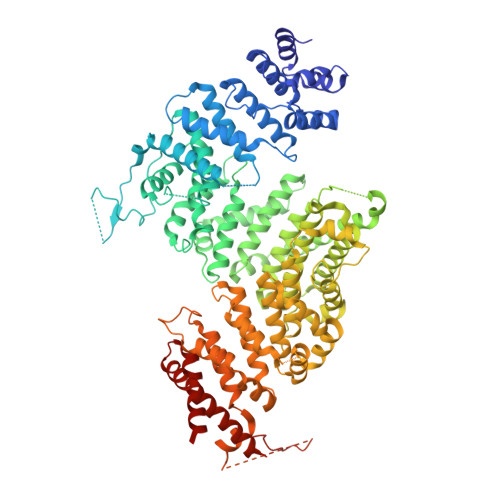
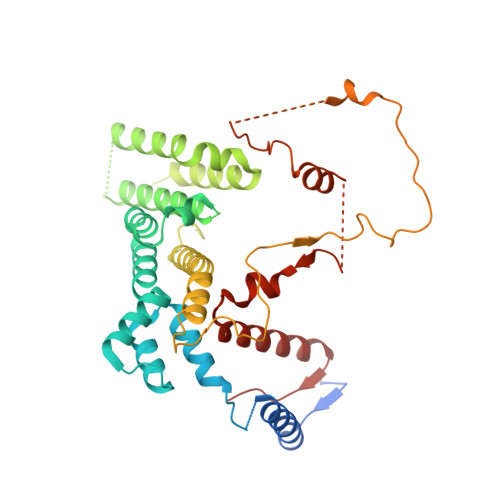
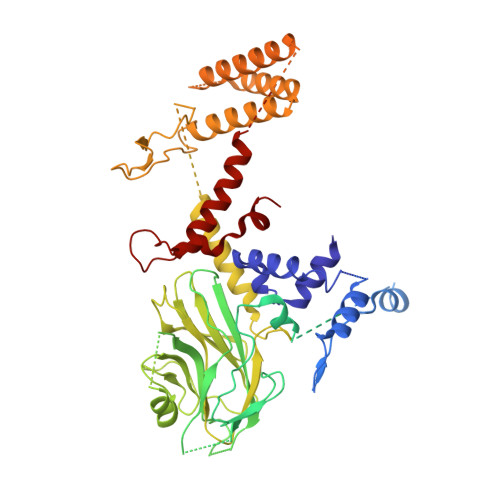
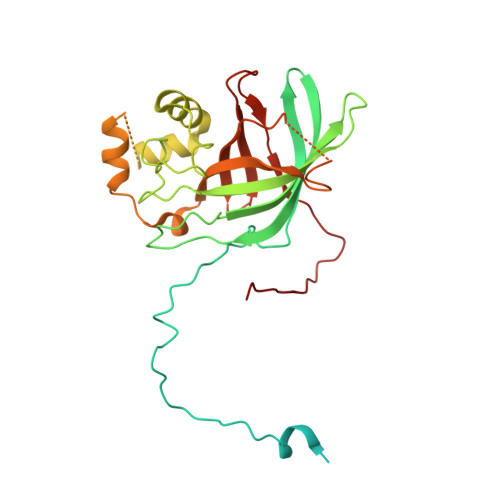
Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Qiao, S., Lee, C.W., Sherpa, D., Chrustowicz, J., Cheng, J., Duennebacke, M., Steigenberger, B., Karayel, O., Vu, D.T., von Gronau, S., Mann, M., Wilfling, F., Schulman, B.A.(2022) Nat Commun 13: 3041-3041
- PubMed: 35650207
- DOI: https://doi.org/10.1038/s41467-022-30803-9
- Primary Citation of Related Structures:
7WUG - PubMed Abstract:
Protein degradation, a major eukaryotic response to cellular signals, is subject to numerous layers of regulation. In yeast, the evolutionarily conserved GID E3 ligase mediates glucose-induced degradation of fructose-1,6-bisphosphatase (Fbp1), malate dehydrogenase (Mdh2), and other gluconeogenic enzymes. "GID" is a collection of E3 ligase complexes; a core scaffold, RING-type catalytic core, and a supramolecular assembly module together with interchangeable substrate receptors select targets for ubiquitylation. However, knowledge of additional cellular factors directly regulating GID-type E3s remains rudimentary. Here, we structurally and biochemically characterize Gid12 as a modulator of the GID E3 ligase complex. Our collection of cryo-EM reconstructions shows that Gid12 forms an extensive interface sealing the substrate receptor Gid4 onto the scaffold, and remodeling the degron binding site. Gid12 also sterically blocks a recruited Fbp1 or Mdh2 from the ubiquitylation active sites. Our analysis of the role of Gid12 establishes principles that may more generally underlie E3 ligase regulation.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, 82152, Martinsried, Germany.
Organizational Affiliation: