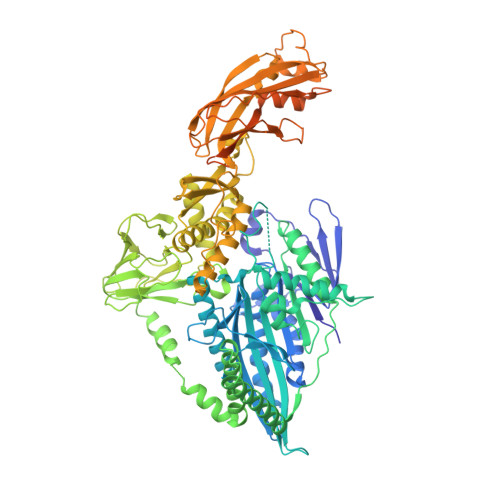
The architecture of kinesin-3 KLP-6 reveals a multilevel-lockdown mechanism for autoinhibition.
Wang, W., Ren, J., Song, W., Zhang, Y., Feng, W.(2022) Nat Commun 13: 4281-4281
- PubMed: 35879313
- DOI: https://doi.org/10.1038/s41467-022-32048-y
- Primary Citation of Related Structures:
7WRG - PubMed Abstract:
Autoinhibition of kinesin-3 ensures the proper spatiotemporal control of the motor activity for intracellular transport, but the underlying mechanism remains elusive. Here, we determine the full-length structure of kinesin-3 KLP-6 in a compact self-folded state. Unexpectedly, all the internal coiled-coil segments and domains in KLP-6 cooperate to successively lock down the neck and motor domains. The first coiled-coil segment is melted into several short helices that work with the motor domain to restrain the entire neck domain. The second coiled-coil segment associates with its neighboring FHA and MBS domains and integrates with the tail MATH domain to form a supramodule that synergistically wraps around the motor domain to trap the nucleotide and hinder the microtubule binding. This multilevel-lockdown mechanism for autoinhibition could be applicable to other kinesin-3 motors.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, 100101, Beijing, China.
Organizational Affiliation: