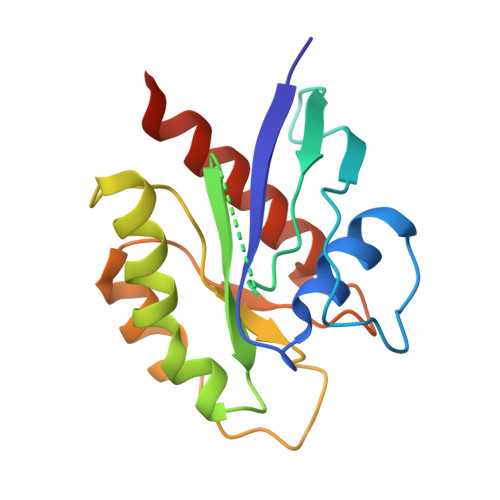
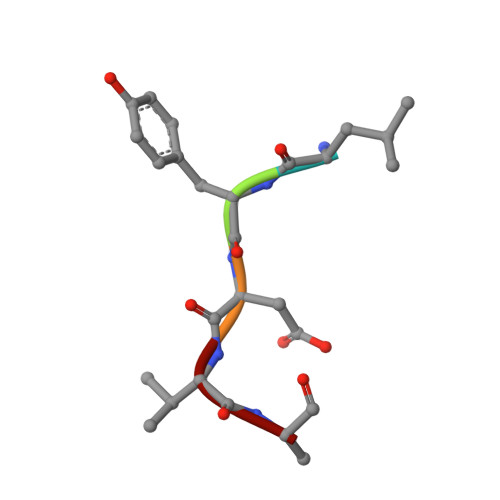
Structural basis of the oncogenic KRAS mutant and GJ101 complex.
Kim, H.J., Han, C.W., Jeong, M.S., Jang, S.B.(2023) Biochem Biophys Res Commun 641: 27-33
- PubMed: 36516586
- DOI: https://doi.org/10.1016/j.bbrc.2022.12.019
- Primary Citation of Related Structures:
7W5R - PubMed Abstract:
KRAS mutations occur in a quarter of all human cancers. When activated in its GTP-bound form, RAS stimulates diverse cellular systems, such as cell division, differentiation, growth, and apoptosis through the activations of various signaling pathways, which include mitogen-activated protein kinase (MAPK), phosphoinositide 3 kinases (PI3K), and RAL-GEFs pathways. We found that GJ101 ( 65 LYDVA 69 ) binds directly to the KRAS mutant (G12V) and showed tumor-suppressive activity. In addition, the GJ101 peptide inhibited KRAS mutant as determined by a [α- 32 P] guanosine triphosphate (GTP) binding assay and suppressed pancreatic cell line in a cell proliferation assay. Herein, the complex structure of KRAS and GJ101 was clarified by X-ray crystallography. Isothermal titration calorimetry showed that GJ101 binds highly with KRAS mutant and the complex structure of KRAS G12V . GJ101 complex presented that the residue of Q61 directly interacted with L65 of GJ101. Overall, the results suggest GJ101 be considered a developmental starting point for KRAS G12V inhibitor.
- Department of Molecular Biology, College of Natural Sciences, Pusan National University, 2, Busandaehak-ro 63beon-gil, Geumjeong-gu, Busan, 46241, Republic of Korea.
Organizational Affiliation: