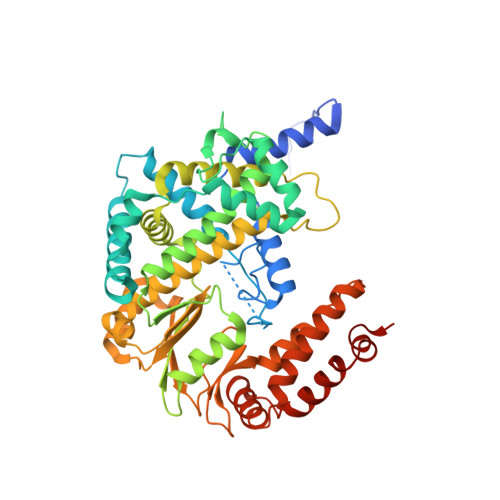
Crystal structures of alphavirus nonstructural protein 4 (nsP4) reveal an intrinsically dynamic RNA-dependent RNA polymerase fold.
Tan, Y.B., Lello, L.S., Liu, X., Law, Y.S., Kang, C., Lescar, J., Zheng, J., Merits, A., Luo, D.(2022) Nucleic Acids Res 50: 1000-1016
- PubMed: 35037043
- DOI: https://doi.org/10.1093/nar/gkab1302
- Primary Citation of Related Structures:
7F0S, 7VB4, 7VW5 - PubMed Abstract:
Alphaviruses such as Ross River virus (RRV), chikungunya virus (CHIKV), Sindbis virus (SINV), and Venezuelan equine encephalitis virus (VEEV) are mosquito-borne pathogens that can cause arthritis or encephalitis diseases. Nonstructural protein 4 (nsP4) of alphaviruses possesses RNA-dependent RNA polymerase (RdRp) activity essential for viral RNA replication. No 3D structure has been available for nsP4 of any alphaviruses despite its importance for understanding alphaviral RNA replication and for the design of antiviral drugs. Here, we report crystal structures of the RdRp domain of nsP4 from both RRV and SINV determined at resolutions of 2.6 Å and 1.9 Å. The structure of the alphavirus RdRp domain appears most closely related to RdRps from pestiviruses, noroviruses, and picornaviruses. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) and nuclear magnetic resonance (NMR) methods showed that in solution, nsP4 is highly dynamic with an intrinsically disordered N-terminal domain. Both full-length nsP4 and the RdRp domain were capable to catalyze RNA polymerization. Structure-guided mutagenesis using a trans-replicase system identified nsP4 regions critical for viral RNA replication.
- Lee Kong Chian School of Medicine, Nanyang Technological University, 59 Nanyang Drive, Singapore 636921.
Organizational Affiliation: